What is Vinculin?
Vinculin is a protein that couples, transmits, transduces, and regulates mechanical force between the cytoskeleton and adhesion receptors (reviewed in [1]).
Vinculin frequently links adhesion receptors (e.g. integrins) to the contractile actin-myosin cytoskeleton by binding either talin through its amino-terminal globular head domain [2], or paxillin through its rod-like tail domain [3]. Vinculin can also bind to lipids through the tail domain. The head and tail domains are linked by a flexible hinge that also contains binding sites for components of the actin polymerizing module (e.g. Arp2/3 complex [4], Ena/VASP proteins [5][6]).
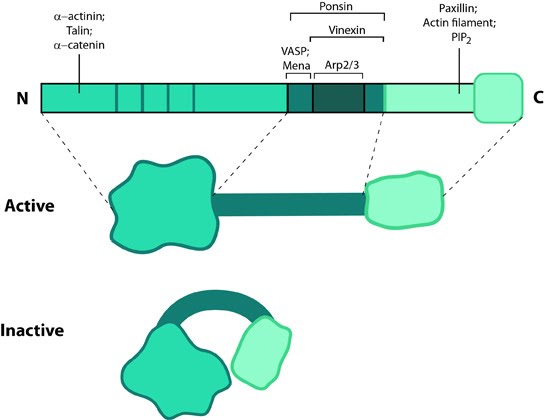
This schematic diagram illustrates the molecular organization of vinculin and shows how vinculin is represented throughout this resource. Relevant domains believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [16893648])
Vinculin generally forms two structural states, an open (active) and closed (inactive) state, which are controlled by interaction(s) between the head and tail domains [7][8]. Whether vinculin can bind to other factors depends both allosterically and sterically on the formation of the complete open state (reviewed in [9]). This in turn is favored by combinatorial binding of ligands namely talin, phosphatidylinositol 4,5-bis-phosphate [PIP2] and actin [8][10][11][12](reviewed in [1]). Phosphorylation at 4 residues has been proposed to prime vinculin for this complex formation and hence the activation process [13]. Activation of vinculin influences its ability to form oligomers or other complxes in cells [14][15].
Localization of Vinculin
Vinculin is present in all adherens junctions [16]. Lipid binding regulates its placement in the adhesions as well as disassembly hence stimulating motility (reviewed in [9]).
Vinculin recruitment to adhesion sites is mechanically regulated by ligand binding(e.g. other cells, extracellullar matrix) and activation of adhesion receptors. Physical restructuring of adhesion receptors such as integrins and their linked mechanosensors (e.g. talin) is transmitted to the cell interior to stimulate contraction of actin stress fibers; this promotes vinculin binding to these sites [2] and subsequent ordering of vinculin domains (reviewed in [8]). Interestingly, reduced cellular tension doesn’t lead to altered vinculin binding as has been observed for other structural components (e.g. zyxin [17]).
Functions of Vinculin
Vinculin N-terminal, when bound to talin, partially opens up and aids integrin clustering for FA growth, possibly by retaining the activated state of integrins [18][19]. The C-terminal forms a mechanosensitive link between adhesion receptors and the actin cytoskeleton to help recruit other components of the actin linking module (e.g. α-actinin, paxillin) and influence the mechanical strength of the cell [2][19][20]. Additionally, vinculin possesses actin filament capping activity [21]. This needs an complete opening of vinculin structure allowing C-terminal of the tail to compete with formin mDia1 for actin barbed ends.
Vinculin also contributes to stability of focal adhesion under high forces by regulating contractile stress generation [2], thereby influencing the cell migration speed [22]. Cells deficient in vinculin cannot form lamellipodia, assemble stress fibers, or spread efficiently over a substrate [19].
References
- Mierke CT. The role of vinculin in the regulation of the mechanical properties of cells. Cell Biochem. Biophys. 2009; 53(3):115-26. [PMID: 19350419]
- Mierke CT, Kollmannsberger P, Zitterbart DP, Smith J, Fabry B, and Goldmann WH. Mechano-coupling and regulation of contractility by the vinculin tail domain. Biophys. J. 2007; 94(2):661-70. [PMID: 17890382]
- Turner CE, Glenney JR, and Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. J. Cell Biol. 1990; 111(3):1059-68. [PMID: 2118142]
- DeMali KA, Barlow CA, and Burridge K. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J. Cell Biol. 2002; 159(5):881-91. [PMID: 12473693]
- Reinhard M, Rüdiger M, Jockusch BM, and Walter U. VASP interaction with vinculin: a recurring theme of interactions with proline-rich motifs. FEBS Lett. 1996; 399(1-2):103-7. [PMID: 8980130]
- Brindle NP, Holt MR, Davies JE, Price CJ, and Critchley DR. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem. J. 1996; 318 ( Pt 3):753-7. [PMID: 8836115]
- Johnson RP, and Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature 1995; 373(6511):261-4. [PMID: 7816144]
- Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, and Liddington RC. Structural basis for vinculin activation at sites of cell adhesion. Nature 2004; 430(6999):583-6. [PMID: 15195105]
- Ziegler WH, Liddington RC, and Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006; 16(9):453-60. [PMID: 16893648]
- Weekes J, Barry ST, and Critchley DR. Acidic phospholipids inhibit the intramolecular association between the N- and C-terminal regions of vinculin, exposing actin-binding and protein kinase C phosphorylation sites. Biochem. J. 1996; 314 ( Pt 3):827-32. [PMID: 8615776]
- Chen H, Choudhury DM, and Craig SW. Coincidence of actin filaments and talin is required to activate vinculin. J. Biol. Chem. 2006; 281(52):40389-98. [PMID: 17074767]
- Cohen DM, Kutscher B, Chen H, Murphy DB, and Craig SW. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J. Biol. Chem. 2006; 281(23):16006-15. [PMID: 16608855]
- Golji J, Wendorff T, and Mofrad MRK. Phosphorylation primes vinculin for activation. Biophys. J. 2012; 102(9):2022-30. [PMID: 22824265]
- Molony L, and Burridge K. Molecular shape and self-association of vinculin and metavinculin. J. Cell. Biochem. 1985; 29(1):31-6. [PMID: 3932372]
- Hüttelmaier S, Mayboroda O, Harbeck B, Jarchau T, Jockusch BM, and Rüdiger M. The interaction of the cell-contact proteins VASP and vinculin is regulated by phosphatidylinositol-4,5-bisphosphate. Curr. Biol. 1998; 8(9):479-88. [PMID: 9560340]
- Geiger B, Volk T, and Volberg T. Molecular heterogeneity of adherens junctions. J. Cell Biol. 1985; 101(4):1523-31. [PMID: 3930512]
- Lele TP, Pendse J, Kumar S, Salanga M, Karavitis J, and Ingber DE. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J. Cell. Physiol. 2006; 207(1):187-94. [PMID: 16288479]
- Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, and Ballestrem C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 2007; 179(5):1043-57. [PMID: 18056416]
- Ezzell RM, Goldmann WH, Wang N, Parashurama N, Parasharama N, and Ingber DE. Vinculin promotes cell spreading by mechanically coupling integrins to the cytoskeleton. Exp. Cell Res. 1997; 231(1):14-26. [PMID: 9056408]
- Alenghat FJ, Fabry B, Tsai KY, Goldmann WH, and Ingber DE. Analysis of cell mechanics in single vinculin-deficient cells using a magnetic tweezer. Biochem. Biophys. Res. Commun. 2000; 277(1):93-9. [PMID: 11027646]
- Le Clainche C, Dwivedi SP, Didry D, and Carlier M. Vinculin is a dually regulated actin filament barbed end-capping and side-binding protein. J. Biol. Chem. 2010; 285(30):23420-32. [PMID: 20484056]
- Goldmann WH, Schindl M, Cardozo TJ, and Ezzell RM. Motility of vinculin-deficient F9 embryonic carcinoma cells analyzed by video, laser confocal, and reflection interference contrast microscopy. Exp. Cell Res. 1995; 221(2):311-9. [PMID: 7493629]


