What is talin?
Talin contains a 47-kDa N-terminal head, a neck and a 220kDa rod domain. The head domain comprises four subdomains termed F0, F1, F2 and F3, with the latter three forming a three-lobed FERM domain.
Integrin tail binding occurs via the F3 phosphotyrosine binding (PTB) domain via a unique interaction with the integrin membrane proximal region, which is sufficient for integrin activation [1]. The basic patches on all subdomains can dock onto the plasma membrane and further enhance integrin activation. Specific interactions through basic residues on F3 are also essential for integrin clustering [2].
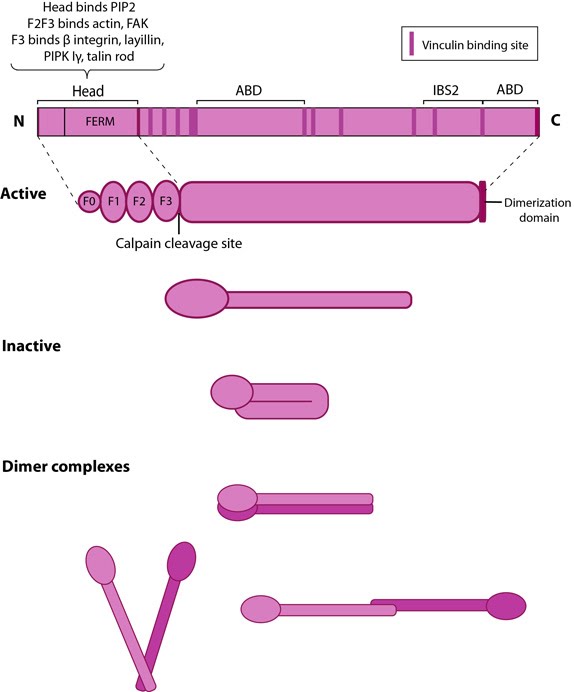
This schematic diagram illustrates the molecular organization of talin (reviewed in [6, 7, 18]) and shows how talin is represented throughout this resource. Hypothetical dimer complexes are presented [19]. ABD = actin binding domain, IBS2 = integrin binding site 2, PIP2 = phosphatidylinositol-4,5-bisphosphate, FAK = focal adhesion kinase, PIPK = phosphatidylinositol phosphate type 1γ.
The rod contains an additional integrin-binding site (IBS2), two actin-binding sites (ABD) and several vinculin-binding sites that are shown to be exposed by stretch in response to force, both in vitro [6][7][8] and in vivo [9]. Vinculin binding reinforces and increases the stability of adhesion sites [10]. Talin also contains numerous potential phosphorylation sites [11] which are suggested to directly or indirectly regulate the association of talin with other factors (reviewed in [4]).
Talin activation and membrane recruitment
Talin exists in an autoinhibited form in the cytosol due to the intermolecular association between the F3 subdomain and a helical bundle in the rod region [12][13]. This not only blocks integrin binding site on F3 but also F2 and F3 binding to membrane. Activation predominantly occurs inside FAs [14][15] and likely involves binding to membrane phospholipids such as phosphatidylinositol 4,5-bis-phosphate (PIP2) [16][17] (reviewed in [18]), vinculin and F-actin [14] or calpain cleavage [5]. This enhances talin’s affinity for the β-integrin subunit by revealing binding sites. Agonist stimulation has been shown to trigger a signaling pathway for membrane targeting of talin/activation of integrin αIIbβ3 [19], involving small GTPase Rap1, Rap-GEF or protein kinase C and adaptor protein RIAM [20][21].
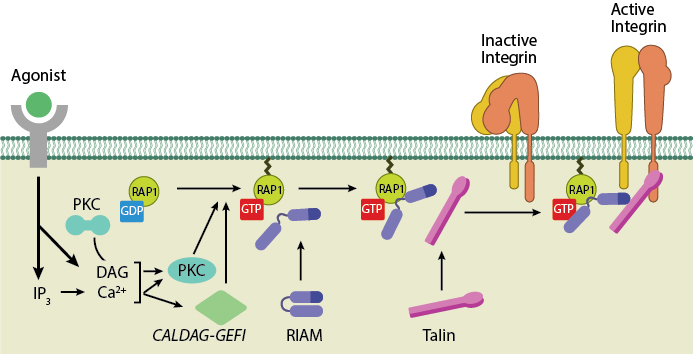
Talin recruitment to membrane. Ligand occupancy in certain cell-surface receptors (agonists) causes phospholipid hydrolysis releasing diacylglycerol (DAG) and inositol triphosphate (IP3). IP3 increases cytosolic levels of calcium ions; DAG and Ca2+ can promote GTP-loading and membrane translocation of Rap1 either by activating Ca2+ and DAG-regulated GEF (CALDAG-GEF or Rap-GEF) or protein kinase C (PKC). Activated Rap1 in turn, recruits Rap1-GTP-interacting adaptor molecule (RIAM) along with its binding partner, talin to the plasma membrane. Adapted from [18573917, 20308986].
Talin is abundant specifically at sites of cell-ECM linkage [22] where it appears to be a key endpoint for multiple signaling pathways that lead to integrin activation (reviewed in [23]). Talin behaves as a prominent structural platform that is required for the initial linkage between the contractile cytoskeleton and sites of integrin/fibronectin adhesion [24].
During cell spreading, talin undergoes cycles of stretching and vinculin binding due to contractile forces on the rearward moving actin filaments [25]. This phenomenon serves to convert matrix forces into biochemical signals at the adhesion site. Hence it not only organizes and stabilizes these initial linkages [10], but it also mediates signal transduction events through the integrins, vinculin and actin (reviewed in [4][18][26]).
The proteolytic cleavage of talin has been shown to be a critical event in the subsequent disassembly of other focal adhesion components [27] but not in integrin activation. Although talin is a key factor that translates mechanical forces into chemical responses primarily at sites of cell-matrix and cell-cell junctions, talin may also function in other cellular processes including membrane ruffling, cytokinesis, and phagocytosis (reviewed in [4]).
References
- Calderwood DA, Yan B, de Pereda JM, Alvarez BG, Fujioka Y, Liddington RC, and Ginsberg MH. The phosphotyrosine binding-like domain of talin activates integrins. J. Biol. Chem. 2002; 277(24):21749-58. [PMID: 11932255]
- Saltel F, Mortier E, Hytönen VP, Jacquier M, Zimmermann P, Vogel V, Liu W, and Wehrle-Haller B. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J. Cell Biol. 2009; 187(5):715-31. [PMID: 19948488]
- Lee H, Bellin RM, Walker DL, Patel B, Powers P, Liu H, Garcia-Alvarez B, de Pereda JM, Liddington RC, Volkmann N, Hanein D, Critchley DR, and Robson RM. Characterization of an actin-binding site within the talin FERM domain. J. Mol. Biol. 2004; 343(3):771-84. [PMID: 15465061]
- Critchley DR. Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem. Soc. Trans. 2004; 32(Pt 5):831-6. [PMID: 15494027]
- Yan B, Calderwood DA, Yaspan B, and Ginsberg MH. Calpain cleavage promotes talin binding to the beta 3 integrin cytoplasmic domain. J. Biol. Chem. 2001; 276(30):28164-70. [PMID: 11382782]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, and Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science 2009; 323(5914):638-41. [PMID: 19179532]
- Lee SE, Kamm RD, and Mofrad MRK. Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J Biomech 2007; 40(9):2096-106. [PMID: 17544431]
- Hytönen VP, and Vogel V. How force might activate talin’s vinculin binding sites: SMD reveals a structural mechanism. PLoS Comput. Biol. 2008; 4(2):e24. [PMID: 18282082]
- Margadant F, Chew LL, Hu X, Yu H, Bate N, Zhang X, and Sheetz M. Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol. 2011; 9(12):e1001223. [PMID: 22205879]
- Jiang G, Giannone G, Critchley DR, Fukumoto E, and Sheetz MP. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 2003; 424(6946):334-7. [PMID: 12867986]
- Ratnikov B, Ptak C, Han J, Shabanowitz J, Hunt DF, and Ginsberg MH. Talin phosphorylation sites mapped by mass spectrometry. J. Cell. Sci. 2005; 118(Pt 21):4921-3. [PMID: 16254238]
- Goksoy E, Ma Y, Wang X, Kong X, Perera D, Plow EF, and Qin J. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol. Cell 2008; 31(1):124-33. [PMID: 18614051]
- Goult BT, Bate N, Anthis NJ, Wegener KL, Gingras AR, Patel B, Barsukov IL, Campbell ID, Roberts GCK, and Critchley DR. The structure of an interdomain complex that regulates talin activity. J. Biol. Chem. 2009; 284(22):15097-106. [PMID: 19297334]
- Banno A, Goult BT, Lee H, Bate N, Critchley DR, and Ginsberg MH. Subcellular localization of talin is regulated by inter-domain interactions. J. Biol. Chem. 2012; 287(17):13799-812. [PMID: 22351767]
- Rossier O, Octeau V, Sibarita J, Leduc C, Tessier B, Nair D, Gatterdam V, Destaing O, Albigès-Rizo C, Tampé R, Cognet L, Choquet D, Lounis B, and Giannone G. Integrins β1 and β3 exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nat. Cell Biol. 2012; 14(10):1057-67. [PMID: 23023225]
- Gilmore AP, and Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature 1996; 381(6582):531-5. [PMID: 8632828]
- Martel V, Racaud-Sultan C, Dupe S, Marie C, Paulhe F, Galmiche A, Block MR, and Albiges-Rizo C. Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J. Biol. Chem. 2001; 276(24):21217-27. [PMID: 11279249]
- Roberts GCK, and Critchley DR. Structural and biophysical properties of the integrin-associated cytoskeletal protein talin. Biophys Rev 2009; 1(2):61-69. [PMID: 19655048]
- Watanabe N, Bodin L, Pandey M, Krause M, Coughlin S, Boussiotis VA, Ginsberg MH, and Shattil SJ. Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin alphaIIbbeta3. J. Cell Biol. 2008; 181(7):1211-22. [PMID: 18573917]
- Bos JL. Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 2005; 17(2):123-8. [PMID: 15780587]
- Boettner B, and Van Aelst L. Control of cell adhesion dynamics by Rap1 signaling. Curr. Opin. Cell Biol. 2009; 21(5):684-93. [PMID: 19615876]
- Geiger B, Volk T, and Volberg T. Molecular heterogeneity of adherens junctions. J. Cell Biol. 1985; 101(4):1523-31. [PMID: 3930512]
- Nayal A, Webb DJ, and Horwitz AF. Talin: an emerging focal point of adhesion dynamics. Curr. Opin. Cell Biol. 2004; 16(1):94-8. [PMID: 15037311]
- DePasquale JA, and Izzard CS. Accumulation of talin in nodes at the edge of the lamellipodium and separate incorporation into adhesion plaques at focal contacts in fibroblasts. J. Cell Biol. 1991; 113(6):1351-9. [PMID: 1904445]
- Smith A, Sengupta K, Goennenwein S, Seifert U, and Sackmann E. Force-induced growth of adhesion domains is controlled by receptor mobility. Proc. Natl. Acad. Sci. U.S.A. 2008; 105(19):6906-11. [PMID: 18463289]
- Critchley DR, and Gingras AR. Talin at a glance. J. Cell. Sci. 2008; 121(Pt 9):1345-7. [PMID: 18434644]
- Franco SJ, Rodgers MA, Perrin BJ, Han J, Bennin DA, Critchley DR, and Huttenlocher A. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 2004; 6(10):977-83. [PMID: 15448700]


