What is capping protein?
Capping protein is involved in actin filament assembly and disassembly
Capping proteins control access to the free barbed ends of actin filaments and is therefore a major factor affecting actin filament elongation. Capping proteins have a high affinity for barbed ends and their micromolar concentration in the cytoplasm ensures that most barbed ends are capped [1][2]. Depletion of capping protein promotes increased filament assembly away from the leading edge in migrating cells [3]. In vitro experiments reveal that this results in a significant loss of lamellipodia yet a sharp increase in filopodia formation [3].
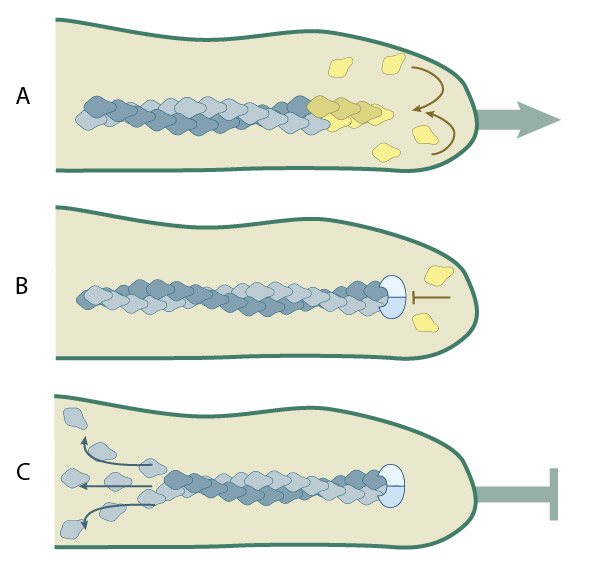
(A) Protrusion of filopodia or lamellipodia is dependent upon actin polymerization at the barbed end of actin filaments. (B) Capping proteins bind to existing actin filaments at the barbed end to prevent filament assembly.(C) ADP-actin continues to depolymerize from the pointed end of the actin filaments. When all of the actin filaments are capped in a filopodium, protrusion is prevented and filament disassembly is favored.
In the case of filopodia formation, the barbed end of a filament can only be extended if the ends of the filament fluctuate away from the membrane to allow the incorporation of new actin monomers; when this occurs, the free barbed ends may become bound by heterodimeric capping proteins [1][2]. Capping proteins are concentrated at the leading edge where they help to modulate the monomeric G-actin^ pool and to control the density and length of actin filaments [4][5] (reviewed in [6]).
Capping proteins (aka β-actinin, CapZ or Cap32/34) help favor actin assembly by preventing the loss of actin subunits to the barbed end (reviewed in [5]). The Ena/VASP protein family accumulates at the plasma membrane to antagonize barbed end capping proteins and to enable actin polymerization into longer filaments [7][8]. Other capping factors (e.g. gelsolin, cytochalasin) bind G-actin monomers and/or unstable oligomers and increase the rate of nucleation [9]. Capping protein activity is inhibited directly by phosphatidylinositol (4,5)-bisphosphate (PIP2), V-1 (aka myotrophin), or CARMIL [1][10][11][12] (reviewed in [5]).
Barbed End Capping
Barbed end capping is essential for efficient disassembly of pointed ends. Capping proteins prevent annealing of severed or debranched filaments and they block both the association and dissociation of subunits at the barbed end [2]. Capping proteins are not only required for profilin to sequester ATP–actin monomers [13], but capping proteins and profilin cooperate to depolymerize filaments faster because of severing and enhanced ADP-actin dissociation from pointed ends [13].
References
- Schafer DA, Jennings PB, and Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 1996; 135(1):169-79. [PMID: 8858171]
- Narita A, Takeda S, Yamashita A, and Maéda Y. Structural basis of actin filament capping at the barbed-end: a cryo-electron microscopy study. EMBO J. 2006; 25(23):5626-33. [PMID: 17110933]
- Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, and Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell 2004; 118(3):363-73. [PMID: 15294161]
- Lai FPL, Szczodrak M, Block J, Faix J, Breitsprecher D, Mannherz HG, Stradal TEB, Dunn GA, Small JV, and Rottner K. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 2008; 27(7):982-92. [PMID: 18309290]
- Iwasa JH, and Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr. Biol. 2007; 17(5):395-406. [PMID: 17331727]
- Wear MA, and Cooper JA. Capping protein: new insights into mechanism and regulation. Trends Biochem. Sci. 2004; 29(8):418-28. [PMID: 15362226]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, and Gertler FB. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 2002; 109(4):509-21. [PMID: 12086607]
- Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, and Faix J. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 2008; 27(22):2943-54. [PMID: 18923426]
- Tellam R, and Frieden C. Cytochalasin D and platelet gelsolin accelerate actin polymer formation. A model for regulation of the extent of actin polymer formation in vivo. Biochemistry 1982; 21(13):3207-14. [PMID: 6285961]
- Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, and Gertler FB. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron 2004; 42(1):37-49. [PMID: 15066263]
- Taoka M, Ichimura T, Wakamiya-Tsuruta A, Kubota Y, Araki T, Obinata T, and Isobe T. V-1, a protein expressed transiently during murine cerebellar development, regulates actin polymerization via interaction with capping protein. J. Biol. Chem. 2002; 278(8):5864-70. [PMID: 12488317]
- Yang C, Pring M, Wear MA, Huang M, Cooper JA, Svitkina TM, and Zigmond SH. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev. Cell 2005; 9(2):209-21. [PMID: 16054028]
- Blanchoin L, Pollard TD, and Mullins RD. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 2000; 10(20):1273-82. [PMID: 11069108]


