What factors influence actin filament length and treadmilling?
Several factors influence actin filament length and treadmilling.
ATP binding on G-actin and free ATP-G-actin concentration
ATP-binding on actin subunits modulates the dynamics of filament assembly, with ATP-binding generally favoring intersubunit interactions and thereby filament assembly [1]. The rate of actin addition to filaments depends on the concentration of free ATP-G-actin whereas the rate of subunit loss does not. At high free ATP-G-actin concentrations the rate of addition exceeds the rate of dissociation and this results in actin filament growth.
The rate of ATP-G-actin assembly to the ends
The addition of ATP-G-actin to the two ends of preexisting actin filaments occurs at very different rates [2]. The addition of free ATP-G-actin at the (-) end is much lower relative to the (+) end.
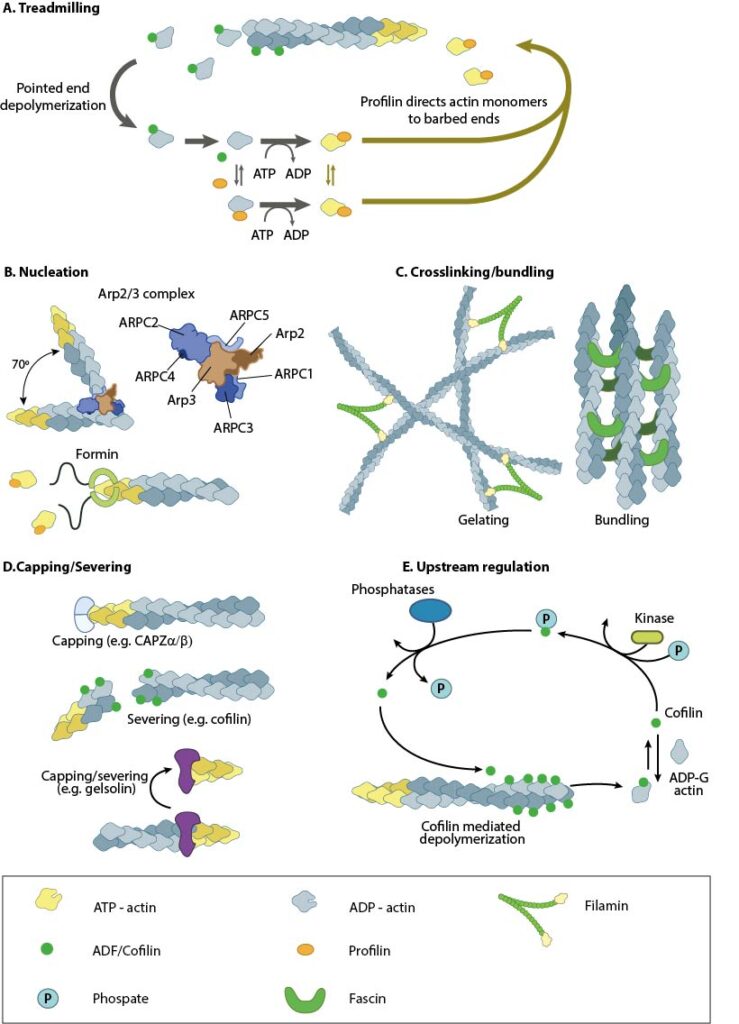
Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.
The critical concentration can be adjusted
The (-) and (+) ends have a different criticial concentration (Cc) for actin filament growth. The Cc is defined as the concentration level of free ATP-G-actin where the rate of addition is balanced by the rate of loss and no net growth occurs at that end. At concentrations above the Cc, actin filament growth occurs, wheras below it, there is a loss of subunits and shrinkage occurs. Any protein that alters the Cc, e.g. profilin, will alter actin filament dynamics.
Actin binding drugs
Toxins such as phalloidins, cytochalasins, latrunculin A, and jasplakinolide are naturally occurring small molecules that bind to actin and alter its polymerization. Phalloidins inhibit actin filament disassembly by locking adjacent actin subunits together, while cytochalasins bind to the barbed end of actin filaments to prevent actin filament assembly and disassembly at that end. Latrunculin A binds to actin monomers to inhibit their polymerization and thus promotes filament disassembly; latrunculin A may also inhibit nucleotide exchange on actin subunits [3][4]. Jasplakinolides stabilize actin monomers, thereby enhancing filament nucleation and assembly.
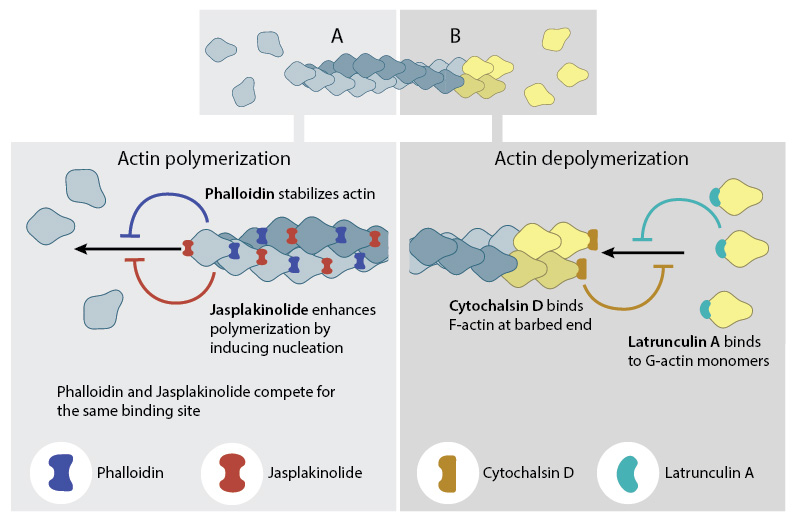
A number of small molecule drugs can bind to actin either during polymerization (A) or depolymerization (B), with varying effects on actin filament dynamics.
Actin binding proteins
Certain actin binding proteins initiate actin filament assembly/disassembly directly, while others influence the ATP binding, the rate of G-actin assembly and the Cc of the filament ends. There are over 60 families of actin-binding proteins (reviewed in [5]) with the main actin monomer-binding proteins in vertebrate cells being thymosin-β4 and profilin. Thymosin-β4 binds strongly to ATP-actin and prevents its assembly into filaments [6]. Because profilin and thymosin-β4 have overlapping binding sites on actin [7][8], profilin must compete with thymosin-β4 during filament assembly [9]. Other examples of actin-binding proteins include: the nucleators spire and the Arp2/3 complex, elongation factors such as formin, actin cross-linking proteins such as fascin or filamin A, nucleation promoting factors such as WASp, and capping protein.
References
- Carlier MF, and Pantaloni D. Control of actin dynamics in cell motility. J. Mol. Biol. 1997; 269(4):459-67. [PMID: 9217250]
- Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 1986; 103(6 Pt 2):2747-54. [PMID: 3793756]
- Yarmola EG, Somasundaram T, Boring TA, Spector I, and Bubb MR. Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A. J. Biol. Chem. 2000; 275(36):28120-7. [PMID: 10859320]
- Morton WM, Ayscough KR, and McLaughlin PJ. Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat. Cell Biol. 2000; 2(6):376-8. [PMID: 10854330]
- Oda T, Stegmann H, Schröder RR, Namba K, and Maéda Y. Modeling of the F-actin structure. Adv. Exp. Med. Biol. 2007; 592:385-401. [PMID: 17278381]
- Carlier MF, Jean C, Rieger KJ, Lenfant M, and Pantaloni D. Modulation of the interaction between G-actin and thymosin beta 4 by the ATP/ADP ratio: possible implication in the regulation of actin dynamics. Proc. Natl. Acad. Sci. U.S.A. 1993; 90(11):5034-8. [PMID: 8506348]
- Safer D, Sosnick TR, and Elzinga M. Thymosin beta 4 binds actin in an extended conformation and contacts both the barbed and pointed ends. Biochemistry 1997; 36(19):5806-16. [PMID: 9153421]
- Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, and Lindberg U. The structure of crystalline profilin-beta-actin. Nature 1993; 365(6449):810-6. [PMID: 8413665]
- Kang F, Purich DL, and Southwick FS. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J. Biol. Chem. 1999; 274(52):36963-72. [PMID: 10601251]


