What are the functions of actin filaments?
Several biological processes related to cell shape and movement depend on actin filaments (reviewed in [1]). Some keys functions are:
- To form the dynamic cytoskeleton, which gives structural support to cells and links the interior of the cell with its surroundings. Forces acting on the actin cytoskeleton are translated and transmitted by signaling pathways to convey information about the external environment.
- To allow cell motility. For example, through the formation and function of Filopodia or Lamellipodia.
- During mitosis, intracellular organelles are transported by motor proteins to the daughter cells along actin cables
- In muscle cells, actin filaments are aligned and myosin proteins generate forces on the filaments to support muscle contraction. These complexes are known as ‘thin filaments’.
- In non-muscle cells, actin filaments form a track system for cargo transport that is powered by non-conventional myosins such as myosin V and VI. Non-conventional myosins use the energy from ATP hydrolysis to transport cargo (such as vesicles and organelles) at rates much faster than diffusion.
Thin filament
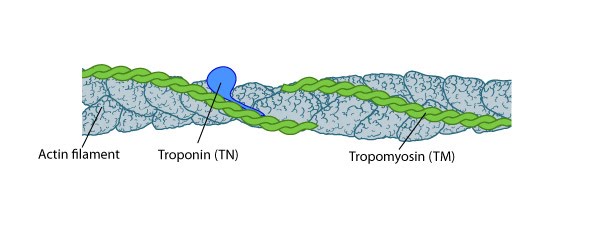
TM binds to the side of adjacent actin subunits along the groove of the helix to stabilize and stiffen the actin filament [4894870]. TM also prevents other proteins from accessing the filament; this inhibition is essential for regulating muscle contraction [17178912]. TN controls the positioning of TM along the groove of the actin filament.
A single tropomyosin binds to the side of adjacent actin subunits and extends over approximately seven actin monomers [2]. End-to-end binding of tropomyosins produces a continuous strand of tropomyosin polymers along the groove of the actin helix which allows their cooperative movement [3]. Tropomyosin isoforms stabilize actin filaments [4] and occupy the same binding sites on actin that are used by known regulators of actin filaments (e.g. ADF/cofilin [5]) (reviewed in [6][7]).
Focus on contractile machinery
Troponin, a three-peptide complex, is thought to trap tropomyosin in a calcium-dependent fashion at a position that inhibits myosin bundles from accessing the actin filaments; calcium binding to troponin allows a conformational restructuring of tropomyosin that leaves the myosin-binding sites on the thin filaments exposed [8][9][10][11]. Subsequent binding of the myosin thick filaments augments movement of tropomyosin away from the actin filament and full exposure of the myosin binding sites [10]. However, control of tropomyosin-binding to myosin thick filaments may be independent of troponin presence [4]; smooth muscle cells and many non-muscle cells lack troponin.
Thus TM regulates both the association of myosin bundles with actin filaments [11] and their ATPase kinetics (reviewed in [12]). It is likely that TM isoforms from different tissues or cell types may have specific effects on actomyosin ATPase activity and cytoskeletal functions [4][13]
References
- Pollard TD, and Cooper JA. Actin, a central player in cell shape and movement. Science 2009; 326(5957):1208-12. [PMID: 19965462]
- Ebashi S, and Endo M. Calcium ion and muscle contraction. Prog. Biophys. Mol. Biol. 1968; 18:123-83. [PMID: 4894870]
- Flicker PF, Phillips GN, and Cohen C. Troponin and its interactions with tropomyosin. An electron microscope study. J. Mol. Biol. 1982; 162(2):495-501. [PMID: 7161805]
- Lehman W, Hatch V, Korman V, Rosol M, Thomas L, Maytum R, Geeves MA, Van Eyk JE, Tobacman LS, and Craig R. Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J. Mol. Biol. 2000; 302(3):593-606. [PMID: 10986121]
- Ono S, and Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 2002; 156(6):1065-76. [PMID: 11901171]
- McGough A. F-actin-binding proteins. Curr. Opin. Struct. Biol. 1998; 8(2):166-76. [PMID: 9631289]
- Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, Bamburg JR, Jeffrey PL, Hardeman EC, Gunning P, and Weinberger RP. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol. Biol. Cell 2003; 14(3):1002-16. [PMID: 12631719]
- Lehman W, Craig R, and Vibert P. Ca(2+)-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature 1994; 368(6466):65-7. [PMID: 8107884]
- Lehman W, Vibert P, Uman P, and Craig R. Steric-blocking by tropomyosin visualized in relaxed vertebrate muscle thin filaments. J. Mol. Biol. 1995; 251(2):191-6. [PMID: 7643394]
- Vibert P, Craig R, and Lehman W. Steric-model for activation of muscle thin filaments. J. Mol. Biol. 1997; 266(1):8-14. [PMID: 9054965]
- Xu C, Craig R, Tobacman L, Horowitz R, and Lehman W. Tropomyosin positions in regulated thin filaments revealed by cryoelectron microscopy. Biophys. J. 1999; 77(2):985-92. [PMID: 10423443]
- Ostap EM. Tropomyosins as discriminators of myosin function. Adv. Exp. Med. Biol. 2008; 644:273-82. [PMID: 19209828]
- Lehman W, and Szent-Györgyi G. Activation of the adenosine triphosphatase of Limulus polyphemus actomyosin by tropomyosin. J. Gen. Physiol. 1972; 59(4):375-87. [PMID: 4260494]


