What are nucleosomes?
In order to fit DNA into the nucleus, it must be packaged into a highly compacted structure known as chromatin. In the first step of this process DNA is condensed into an 11 nm fiber that represents an approximate 6-fold level of compaction [1]. This is achieved through nucleosome assembly.
The nucleosome is the smallest structural component of chromatin, and is produced through interactions between DNA and histone proteins. Here, a histone octamer is formed from the histones H2A, H2B, H3 and H4, although in some cases other histone variants may also be found in the core (e.g., H2A.Z, MacroH2A, H2a.Bbd, H2A.lap1, H2A.X, H3.3, CenH3 and others [1]). A 147bp segment of DNA then wraps around the histone octamer 1.75 times, thus completing the formation of a single nucleosome.
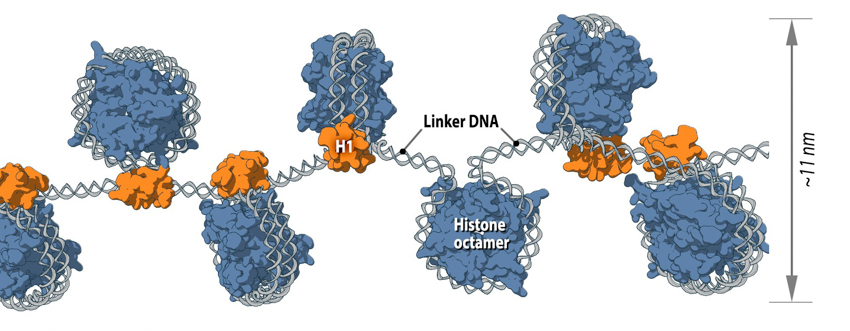
Each nucleosome consists of histone octamer core, assembled from the histones H2A, H2B, H3 and H4 (or other histone variants in some cases) and a segment of DNA that wraps around the histone core. Adjacent nucleosomes are connected via “linker DNA”.
Of course, a single nucleosome will not form in isolation but is instead part of a wider process, whereby multiple nucleosomes form in a linear fashion along the DNA molecule. This ultimately produces the 11 nm fiber, which is traditionally described, based on its appearance, as “beads on a string” [2]. Here, adjacent nucleosomes are connected via “linker DNA”, which is usually bound to the H1 histone and is between 20-80 bps long. Additionally, flexible histone tails which originate from the histone octamer extend away from nucleosomal DNA and can interact with other nucleosomes, stabilizing more complex 3D structures [3]. In other words, specific nucleosomes can be far apart with respect to their linear sequence, but within interacting distance in the context of higher order chromatin structure [1].
Alternative nucleosome conformations (reviewed in [1]) may arise due to spontaneous unwrapping and rewrapping of DNA around the histone core, as well as due to variations in histones themselves. Moreover, nucleosomes are highly dynamic and can undergo spontaneous sliding, “splitting” or even complete dissociation.
The level of compaction attained through the formation of the 11 nm nucleosome fiber is insufficient to package the whole genome into the nucleus. Instead, this fiber forms the basis for other higher order chromatin structures that are established through additional folding and bending events.
Intermediate chromatin structures
Despite the extensive knowledge already gained on the structure of the 11 nm nucleosome fiber, as well as metaphase chromosomes, the intermediate chromatin structures commonly described are largely hypothetical and yet to be observed in vivo.
Two popular models that were proposed based on in vitro data are the solenoid and zigzag. In each case, the 11 nm nucleosome fiber undergoes additional folding to form a 30 nm fiber [4][5] with the manner of folding for a particular region depending on the internucleosomal linker length and the presence of linker histone H17 [6]. In the one-start solenoid model, bent linker DNA sequentially connects each nucleosome cores, creating a structure where nucleosomes follow each other along the same helical path [4][7]. Alternatively, in the two-start zigzag model, straight linker DNA connects two opposing nucleosome cores, creating the opposing rows of nucleosomes that form so called “two-start” helix. In zigzag model, alternate nucleosomes (for example, N1 and N3) become interacting partners [5][8]. Interestingly, some studies offer a model, where intermediate 30 nm fibers contain both the solenoid and zigzag conformations [9], suggesting instead that observations made in in vitro experiments might be an isolation artifact due to strictly cationic low-salt environment or chemical cross-linking (e.g., glutaraldehyde fixation). Consequently, new models of 11 nm fiber compaction have been proposed (e.g., chromonema, chromatin hub, hybrid chromonema/chromatin hub, fractal [10][11][12]), but no common conclusion has been reached yet.
One aspect shared by most of the models for higher order chromatin organization is the dynamic existence of decondensed loops among more compact chromatin structures. In most cases, higher order chromatin has to be decondensed to a nucleosome structural level in order to transcribe genes [13][14]. The length of the decondensed chromatin loop can sometimes exceed the area occupied by the chromosome territory, to which the loop belongs, allowing it to intermingle into the neighbouring chromosome territory [15].
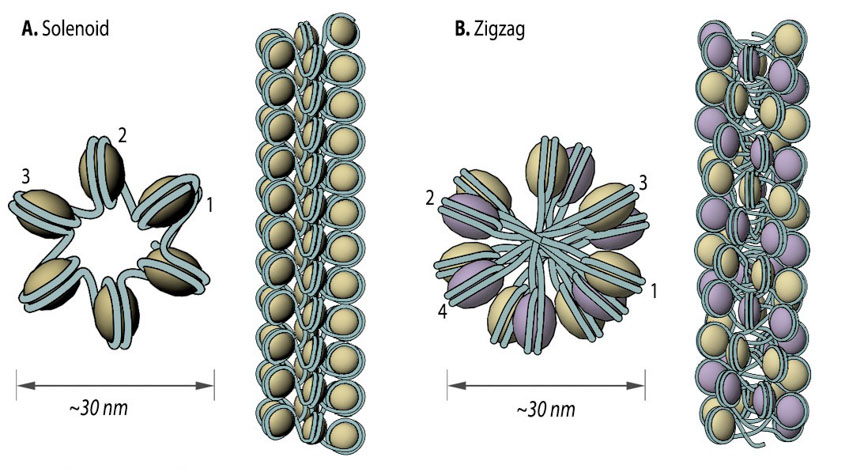
30 nm chromatin fibers are considered to exist in the form of so called solenoid or zigzag. The main feature of solenoid model is that nucleosomes follow each other along the same helical path, and interactions between the histone cores occur sequentially (1, 2, 3 and so on). Therefore, solenoid is also referred to as “one start model”. In zigzag, on the other hand, linker DNA connects two opposing nucleosomes, creating a structure where the alternate histone cores become interacting partners (i.e., 1 and 3, 2 and 4 and so on). Therefore, zigzag is considered as a “two start model”, which is indicated in the figure (B) by two different colors of histone cores: yellow interacting nucleosome partners (1, 3, etc.) as opposed to the violet nucleosome row (2, 4, etc.).
References
- Luger K, Dechassa ML, and Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 2012; 13(7):436-47. [PMID: 22722606]
- Olins DE, and Olins AL. Chromatin history: our view from the bridge. Nat. Rev. Mol. Cell Biol. 2003; 4(10):809-14. [PMID: 14570061]
- Martins RP, Finan JD, Guilak F, and Lee DA. Mechanical regulation of nuclear structure and function. Annu Rev Biomed Eng 2012; 14:431-55. [PMID: 22655599]
- Finch JT, and Klug A. Solenoidal model for superstructure in chromatin. Proc. Natl. Acad. Sci. U.S.A. 1976; 73(6):1897-901. [PMID: 1064861]
- Woodcock CL, Frado LL, and Rattner JB. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J. Cell Biol. 1984; 99(1 Pt 1):42-52. [PMID: 6736132]
- Routh A, Sandin S, and Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. U.S.A. 2008; 105(26):8872-7. [PMID: 18583476]
- Widom J, and Klug A. Structure of the 300A chromatin filament: X-ray diffraction from oriented samples. Cell 1985; 43(1):207-13. [PMID: 4075395]
- Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, and Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 2004; 306(5701):1571-3. [PMID: 15567867]
- Grigoryev SA, Arya G, Correll S, Woodcock CL, and Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl. Acad. Sci. U.S.A. 2009; 106(32):13317-22. [PMID: 19651606]
- Stadhouders R, Thongjuea S, Andrieu-Soler C, Palstra R, Bryne JC, van den Heuvel A, Stevens M, de Boer E, Kockx C, van der Sloot A, van den Hout M, van Ijcken W, Eick D, Lenhard B, Grosveld F, and Soler E. Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J. 2011; 31(4):986-99. [PMID: 22157820]
- Bian Q, and Belmont AS. Revisiting higher-order and large-scale chromatin organization. Curr. Opin. Cell Biol. 2012; 24(3):359-66. [PMID: 22459407]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, and Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326(5950):289-93. [PMID: 19815776]
- Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, and Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 2002; 9(3):541-52. [PMID: 11931762]
- Hu Y, Kireev I, Plutz M, Ashourian N, and Belmont AS. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J. Cell Biol. 2009; 185(1):87-100. [PMID: 19349581]
- Mirny LA. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011; 19(1):37-51. [PMID: 21274616]


