What are chromosomes and chromosome territories?
While metaphase chromosomes can be depicted as distinct bodies with well-defined shapes and sizes, interphase chromosomes are less uniform and, by filling the nuclear space, difficult to distinguish. Despite this, recent research has revealed how the nuclear architecture dictates interphase chromosome arrangement and territorial organization in differentiated cells.
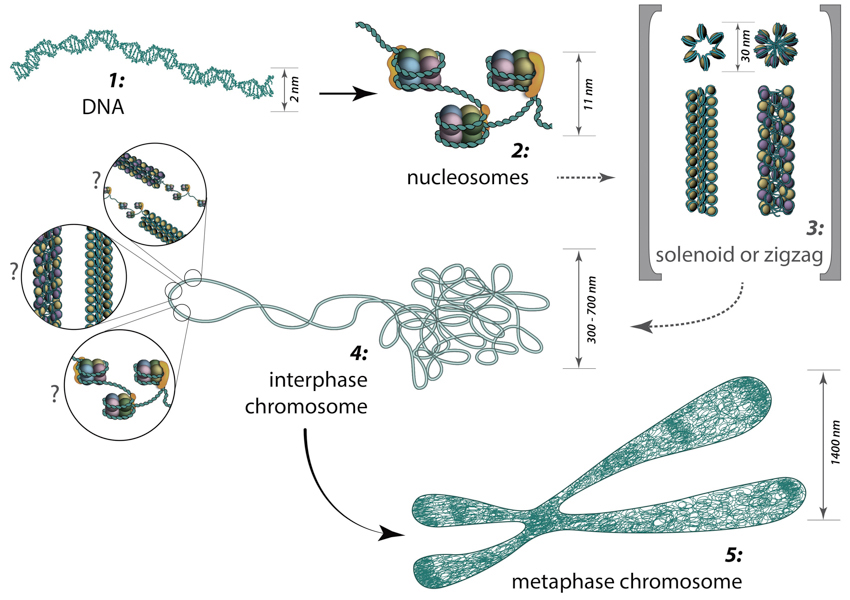
Folding of DNA into nucleosomes achieves initial 6-fold compaction level. Histone variants present in the nucleosome core, posttranslational modifications and linker histone H1 position can all control DNA accessibility for transcription at this compaction level. Further chromatin condensation into 30 nm fibers (i.e., zigzag or solenoid) is suggested by in vitro data and is yet to be proved or discredited to exist in vivo. During the interphase, chromatin is folded into 300-700 nm domains, which together comprise a chromosome territory. The structure and organization of chromatin loops inside a chromosome territory remains the matter of debates and was proposed to exist in the form of solenoid, or zigzag, or nucleosomes, or a hybrid of those.
During interphase, each chromosome occupies a spatially limited, roughly elliptical domain which is known as a chromosome territory (CT) [1][2]. Each chromosome territory is comprised of higher order chromatin units of ~1 Mb each. These units are likely built up from smaller loop domains. On the other hand, 1Mb domains can themselves serve as smaller units in higher-order chromatin structures [1].
Chromosome territories are known to be arranged radially around the nucleus. This arrangement is both cell and tissue-type specific and is also evolutionary conserved [3]. The radial organization of chromosome territories was shown to correlate with their gene density and size. In this case, the gene-rich chromosomes occupy interior positions, whereas larger, gene-poor chromosomes, tend to be located around the periphery [[4][5][6]. Chromosome territories are also dynamic structures, with genes able to relocate from the periphery towards the interior once they have been ‘switched on’ [7]. In other cases, genes may move in the opposite direction, or simply maintain their position [8][9]. The eviction of genes from their chromosome territories into the interchromatin compartment or a neighbouring chromosome territory is often accompanied by the formation of large decondensed chromatin loops [3].
Models describing chromosome territory arrangement
With the development of high-throughput biochemical techniques, such as 3C (‘chromosome conformation capture’)[10] and 4C (‘chromosome conformation capture-on-chip’ [11] and ‘circular chromosome conformation capture’ [12]), numerous spatial interactions between neighbouring chromatin territories have been described. These descriptions have been supplemented with the construction of spatial proximity maps for the entire genome (e.g., for a human lymphoblastoid cell line [13]). Together, these observations and physical simulations have led to the proposal of various models that aim to define the structural organization of chromosome territories [1]:
The chromosome territory-interchromatin compartment (CT-IC) model describes two principal compartments: chromosome territories (CTs) and an interchromatin compartment (IC). In this model, chromosome territories build up an interconnected chromatin network [14] that is associated with an adjacent 3D space called the interchromatin compartment. The latter can be observed using both light and electron microscopy [15]
Within a single chromosome territory, the interphase chromosome is divided into defined regions based on the level of chromosome condensation. Here, the inner part of the interphase chromosome is comprised of more condensed chromatin domains or higher-order chromatin fibers, while a thin (<200 nm) layer of more decondensed chromatin, known as the perichromatin region, can be found around the chromosomal periphery [16]. Functionally, the perichromatin region represents the major transcriptional compartment, and is also the region where most co-transcriptional RNA splicing takes place [17][18][19]. DNA replication [20] and DNA repair [21] is also predominately carried out within the perichromatin region. Finally, nascent RNA transcripts, referred to as perichromatin fibrils, are also generated in the perichromatin region. Perichromatin fibrils are then subjected to the splicing events by the factors, provided from the interchromatin compartment.
The lattice model, proposed by Dehgani et al. [22] is based on reports that transcription also occurs within the inner, more condensed chromosome territories and not only at the interface between the interchromatin compartment and the perichromatin region [23][24][25][26]. Using ESI (electron spectroscopic imaging), Dehgani et al. showed that chromatin was organized as an array of deoxy-ribonucleoprotein fibers of 10–30 nm in diameter. In this study, the interchromatin compartments, which are described in the CT-IC model as large channels between chromosome territories, were not apparent. Instead, chromatin fibers created a loose meshwork of chromatin throughout the nucleus that intermingled at the periphery of chromosome territories. Thus, inter- and intra-chromosomal spaces within this meshwork are essentially contiguous and together form the intra-nuclear space [22].
The interchromatin network (ICN) model [27] predicts that intermingling chromatin fibers/loops can make both cis- (within the same chromosome) and trans- (between different chromosomes) contacts. This intermingling is uniform and makes distinction between the chromosome territory and interchromatin compartment functionally meaningless [1]. The advantage of the ICN model is that it permits high chromatin dynamics and diffusion-like movements. The authors propose that ongoing transcription influences the degree of intermingling between specific chromosomes by stabilizing associations between particular loci. Such interactions are likely to depend on the transcriptional activity of the loci, and are therefore cell-type specific.
The cell type-specific organization of chromosome territories has been studied by measuring the volume and frequency of intermingling between heterologous chromosomes. By using 3C (chromosome conformation capture) and FISH (fluorescence in situ hybridization) to map the regions of chromosome intermingling, it was revealed that these regions contain a higher density of active genes and are enriched with markers of transcriptional activation and repression, such as activated RNAPII. By comparing the positions of the CTs in undifferentiated mouse embryonic stem (ES) cells, ES cells in early stages of differentiation, and terminally differentiated NIH3T3 cells, it was shown that fully differentiated cells had a higher enrichment of RNAPII, compared to undifferentiated or less-differentiated cells. The findings support the notion that the intermingling regions have functional significance in the nucleus and provide a basis for understanding how the radial and relative positions of chromosomal territories evolve during the process of differentiation, explaining their organization in a cell type-dependent manner [28].
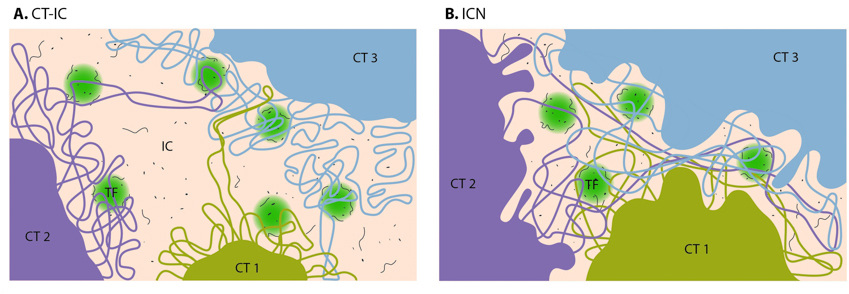
On the CT-IC model, the space between discrete CTs can be visualized in light and electron microscope and is called interchromatin compartment (IC). Transcription factories (TF, green color) are localized predominantly in perichromatin region. In the ICN model, interchromatin compartment is not apparent. Instead, the space between CTs is occupied by intermingling decondensed chromatin loops, which often share the same transcription factories.
The Fraser and Bickmore model [29] emphasizes the functional importance of giant chromatin loops, which originate from chromosome territories and expand across the nuclear space in order to share transcription factories. In this case, both cis- and trans- loops of decondensed chromatin can be co-expressed and co-regulated by the same transcription factory.
The Chromatin polymer models assume a broad range of chromatin loop sizes [30] and predict the observed distances between genomic loci and chromosome territories, as well as the probabilities of contacts being formed between given loci [31]. These models apply physics-based approaches that highlight the importance of entropy for understanding nuclear organization. By proposing the existence of conformational chromatin ensembles with structures based on three possible homopolymer states, these models also provide alternative structures to the traditional 30 nm chromatin fiber, which has been brought into question following recent studies [32][33][34].
With a lack of experimental evidence to support these described models, it must be remembered that they serve only to hypothesize the structural and chemical properties of intermediate chromatin structures, and to highlight unanswered questions [1]. For example, the mechanisms that exist to control the rate and the extent of chromatin movement remain to be defined.
References
- Cremer T, and Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol 2010; 2(3):a003889. [PMID: 20300217]
- Khalil A, Grant JL, Caddle LB, Atzema E, Mills KD, and Arneodo A. Chromosome territories have a highly nonspherical morphology and nonrandom positioning. Chromosome Res. 2007; 15(7):899-916. [PMID: 17926137]
- Meaburn KJ, and Misteli T. Cell biology: chromosome territories. Nature 2007; 445(7126):379-781. [PMID: 17251970]
- Mayer R, Brero A, von Hase J, Schroeder T, Cremer T, and Dietzel S. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 2005; 6:44. [PMID: 16336643]
- Tanabe H, Habermann FA, Solovei I, Cremer M, and Cremer T. Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat. Res. 2002; 504(1-2):37-45. [PMID: 12106644]
- Sun HB, Shen J, and Yokota H. Size-dependent positioning of human chromosomes in interphase nuclei. Biophys. J. 2000; 79(1):184-90. [PMID: 10866946]
- Chuang C, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, and Belmont AS. Long-range directional movement of an interphase chromosome site. Curr. Biol. 2006; 16(8):825-31. [PMID: 16631592]
- Galiová G, Bártová E, and Kozubek S. Nuclear topography of beta-like globin gene cluster in IL-3-stimulated human leukemic K-562 cells. Blood Cells Mol. Dis. 33(1):4-14. [PMID: 15223004]
- Foster HA, and Bridger JM. The genome and the nucleus: a marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma 2005; 114(4):212-29. [PMID: 16133352]
- Dekker J, Rippe K, Dekker M, and Kleckner N. Capturing chromosome conformation. Science 2002; 295(5558):1306-11. [PMID: 11847345]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, and de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 2006; 38(11):1348-54. [PMID: 17033623]
- Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, and Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006; 38(11):1341-7. [PMID: 17033624]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, and Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326(5950):289-93. [PMID: 19815776]
- Visser AE, Jaunin F, Fakan S, and Aten JA. High resolution analysis of interphase chromosome domains. J. Cell. Sci. 2000; 113 ( Pt 14):2585-93. [PMID: 10862716]
- Rouquette J, Genoud C, Vazquez-Nin GH, Kraus B, Cremer T, and Fakan S. Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: a novel electron-microscopic approach to reconstructing nuclear architecture. Chromosome Res. 2009; 17(6):801-10. [PMID: 19731052]
- Fakan S, and van Driel R. The perichromatin region: a functional compartment in the nucleus that determines large-scale chromatin folding. Semin. Cell Dev. Biol. 2007; 18(5):676-81. [PMID: 17920313]
- Fakan S, and Bernhard W. Localisation of rapidly and slowly labelled nuclear RNA as visualized by high resolution autoradiography. Exp. Cell Res. 1971; 67(1):129-41. [PMID: 4105558]
- Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, Fu XD, van Driel R, and Fakan S. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell 1999; 10(1):211-23. [PMID: 9880337]
- Trentani A, Testillano PS, Risueño MC, and Biggiogera M. Visualization of transcription sites at the electron microscope. Eur J Histochem 2003; 47(3):195-200. [PMID: 14514409]
- Jaunin F, and Fakan S. DNA replication and nuclear architecture. J. Cell. Biochem. 2002; 85(1):1-9. [PMID: 11891845]
- Solimando L, Luijsterburg MS, Vecchio L, Vermeulen W, van Driel R, and Fakan S. Spatial organization of nucleotide excision repair proteins after UV-induced DNA damage in the human cell nucleus. J. Cell. Sci. 2008; 122(Pt 1):83-91. [PMID: 19066286]
- Dehghani H, Dellaire G, and Bazett-Jones DP. Organization of chromatin in the interphase mammalian cell. Micron 2005; 36(2):95-108. [PMID: 15629642]
- Visser AE, Eils R, Jauch A, Little G, Bakker PJ, Cremer T, and Aten JA. Spatial distributions of early and late replicating chromatin in interphase chromosome territories. Exp. Cell Res. 1998; 243(2):398-407. [PMID: 9743599]
- Verschure PJ, van der Kraan I, Manders EMM, Hoogstraten D, Houtsmuller AB, and van Driel R. Condensed chromatin domains in the mammalian nucleus are accessible to large macromolecules. EMBO Rep. 2003; 4(9):861-6. [PMID: 12947417]
- Verschure PJ, van Der Kraan I, Manders EM, and van Driel R. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 1999; 147(1):13-24. [PMID: 10508851]
- Mahy NL, Perry PE, Gilchrist S, Baldock RA, and Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 2002; 157(4):579-89. [PMID: 11994314]
- Branco MR, and Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006; 4(5):e138. [PMID: 16623600]
- Maharana S, Iyer KV, Jain N, Nagarajan M, Wang Y, and Shivashankar GV. Chromosome intermingling-the physical basis of chromosome organization in differentiated cells. Nucleic Acids Res. 2016; 44(11):5148-60. [PMID: 26939888]
- Fraser P, and Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature 2007; 447(7143):413-7. [PMID: 17522674]
- Mateos-Langerak J, Bohn M, de Leeuw W, Giromus O, Manders EMM, Verschure PJ, Indemans MHG, Gierman HJ, Heermann DW, van Driel R, and Goetze S. Spatially confined folding of chromatin in the interphase nucleus. Proc. Natl. Acad. Sci. U.S.A. 2009; 106(10):3812-7. [PMID: 19234129]
- Fudenberg G, and Mirny LA. Higher-order chromatin structure: bridging physics and biology. Curr. Opin. Genet. Dev. 2012; 22(2):115-24. [PMID: 22360992]
- Fussner E, Ching RW, and Bazett-Jones DP. Living without 30nm chromatin fibers. Trends Biochem. Sci. 2011; 36(1):1-6. [PMID: 20926298]
- Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, and Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc. Natl. Acad. Sci. U.S.A. 2008; 105(50):19732-7. [PMID: 19064912]
- Maeshima K, and Eltsov M. Packaging the genome: the structure of mitotic chromosomes. J. Biochem. 2007; 143(2):145-53. [PMID: 17981824]


