How is transcription regulated in stem cells?
Pluripotent stem cells can potentially differentiate into any given cell type. They commonly exhibit mechanically softer nuclei [1], lack type-A lamins [2] and have a poorly defined cytoskeleton [3][4].
Embryonic stem cells are pluripotent in early organism development, but gradually undergo lineage restriction and transform into the stem cells with limited differentiation capacities (e.g., hematopoietic stem cells, neural stem cells). These stem cells divide to give rise to specialized cells and tissues, but half of the new generation does not differentiate, retaining their stem cell identity. This type of division cycles is possible due to both epigenetic inheritance (i.e., DNA methylation, chromatin modifiers, histone variants) and chromatin plasticity of stem cells (reviewed in [5]).
Generally, stem cell chromatin is less compact and more transcription-permissive when compared to differentiated cells. Fluorescent recovery after photobleaching (FRAP) experiments showed high exchange rate of several chromatin-associated proteins (e.g., histones H2B and H3, heterochromatin-associated protein HP1 and linker histone H1) in embryonic stem cells [6]. However, a search for molecular “signature” of stem cells failed to retrieve much: only Oct4, Sox2 and Nanog were identified as commonly expressed embryonic stem cell-specific genes (reviewed in [5]). In terms of localization, a region of 12p chromosome that contains clustered pluripotency genes (including Nanog) is confined to more central nuclear regions in human embryonic stem cells as compared to differentiated cells [7]. Furthermore, Oct4 locus was found to loop out from its chromosome territory (chromosome 6p) in human embryonic stem cells [7]. The same study demonstrated that centromeric regions of chromosomes in stem cells are not as much confined to the nuclear periphery as they are in differentiated cells [7].
To describe the transcriptional regulation in embryonic stem cells, Meshorer and Misteli proposed the following models [8]:
- The hierarchical activation (HA) model emphasizes the existence of “stemness” genes, responsible for self-renewal and maintenance of pluripotency. These genes are silenced during cell commitment, while lineage-specific genes are activated.
- The promiscuous transcription (PT) model is similar to HA model in that specific “stemness” genes are expressed; however, many other genes, including lineage-specific, are also indiscriminately expressed at the low “background” level. During cell differentiation, these “background” and “stemness” genes are silenced, and tissue-specific genes are activated.
- The early transcription competence marks (ETCM) model is based on the observation that in embryonic stem cells several tissue-specific genes, albeit repressed, still contain markers of active chromatin (the so-called “bivalent domains” [9]), suggesting that they are marked for expression at a later stage of differentiation.
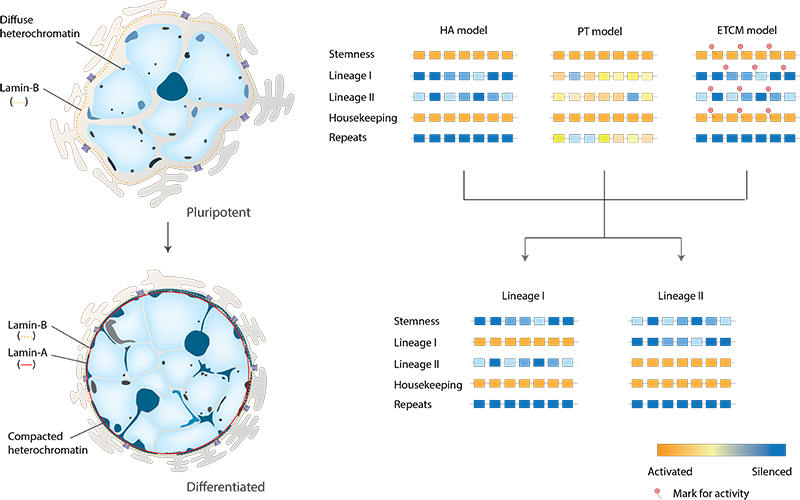
In pluripotent embryonic stem cells, nuclei lack lamin-A and possess a softer nuclear envelope. Undifferentiated embryonic stem cells are richer in less compact euchromatin and diffuse heterochromatin. As cells differentiate, regions of condensed heterochormatin accumulate. The HA model (Hierarchical Activation model); the PT model (Promiscuous Transcription model) and the ETCM model (Early Transcription Competence Marks model) are depicted on the right.
References
- Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, and Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 2007; 104(40):15619-24. [PMID: 17893336]
- Constantinescu D, Gray HL, Sammak PJ, Schatten GP, and Csoka AB. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells 2005; 24(1):177-85. [PMID: 16179429]
- Mazumder A, and Shivashankar GV. Emergence of a prestressed eukaryotic nucleus during cellular differentiation and development. J R Soc Interface 2010; 7 Suppl 3:S321-30. [PMID: 20356876]
- Mazumder A, Roopa T, Kumar A, Iyer KV, Ramdas NM, and Shivashankar GV. Prestressed nuclear organization in living cells. Methods Cell Biol. 2010; 98:221-39. [PMID: 20816237]
- Spivakov M, and Fisher AG. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 2007; 8(4):263-71. [PMID: 17363975]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, and Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 2006; 10(1):105-16. [PMID: 16399082]
- Wiblin AE, Cui W, Clark AJ, and Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J. Cell. Sci. 2005; 118(Pt 17):3861-8. [PMID: 16105879]
- Meshorer E, and Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006; 7(7):540-6. [PMID: 16723974]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, and Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125(2):315-26. [PMID: 16630819]


