How is the nucleus coupled to the cytoskeleton?
Cytoskeletal filaments bridge the nucleus to the plasma membrane, which in turn is anchored at sub-cellular sites to extracellular substrates via a plethora of proteins that form focal adhesions (FAs). FAs are points of cross-talk between transmembrane integrin receptors and the cytoplasmic filaments and thus are key sites for both biochemical and mechanotransduction pathways (reviewed in [1]). Linkage can be direct or via various adaptor proteins, providing structural support to both cellular and nuclear structures (reviewed in [2][3][4])
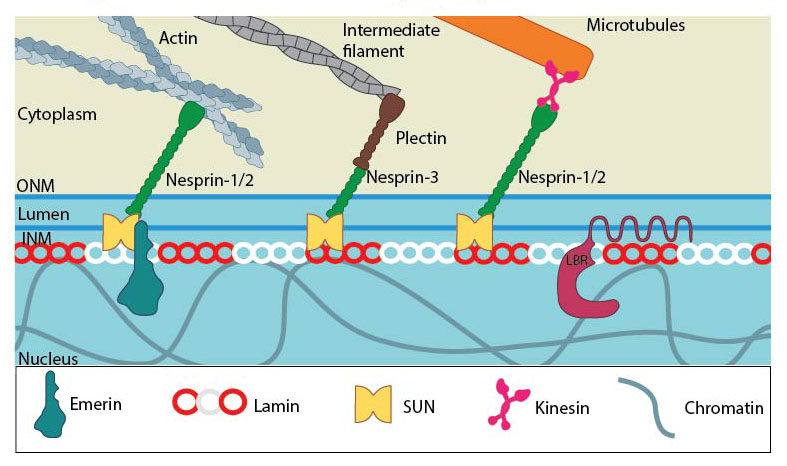
SUN and KASH domain proteins (mainly nesprins) provide the essential linkage between the three main cytoskeletal networks aend the nucleokeleton especially the nuclear lamina. Plectin acts as an intermediary linker connecting the intermediate filaments while a motor protein (e.g: kinesin) is generally involved in linking microtubules. INM- Inner nuclear membrane; ONM- Outer nuclear membrane; LBR- Lamin B receptor. Together these provide the outward pull that maintains the nucleus in a stretched state. Adapted from [21321324, 15688064, 21327104].
These links are emerging to be pivotal in various physiological processes including cell migration, and in the ability for cells to cope with mechanical stress [11][12]. Defects in the complex have serious consequences to nuclear mechanotransduction [13][14]. For example, when the LINC complex in fibroblasts was disrupted by inducing the mislocalization of nesprin-3, aletered gene expression profiles occured, with different sets of genes being over or under expressed depending no whether cells were grown on soft or stiff substrates [15].
How does the cytoskeleton influence nuclear morphology and positioning?
Work by Mazumder et al. ascertained the active involvement of cytoskeletal forces in determining nuclear morphology. Change in nuclear size upon perturbation of actomyosin and microtubules affirmed their roles in exerting tensile and compressive forces respectively on the nucleus, correlating with their functions in the cellular context [16] [17], [18]. Further evidence confirming forces are transmitted to the nucleus from focal adhesions has been demonstrated using fibroblasts plated on microfabricated pillar arrays. Here, traction forces, which were measured by pillar deflection, were positively or negatively, correlated to heterochromatin movement inside the nucleus. In this case deflection of the pillars beneath the leading edge were negatively correlated, whereas those at the trailing edge, and hence in closer proximity to the nucleus, were positively correlated. In this case it was suggested that force propagation occured via an elastic or contractile component of the actin cytoskeleton [19]
Furthermore, the ‘perinuclear cap’, which is composed of contractile actin bundles that bridge focal adhesions on either side of the nucleus, has been shown to tightly regulate the nuclear geometry [20]. These bundles pass apically to form a dome covering the top of the nucleus and are connected to the nucleus through the LINC complexes. They are completely absent in pluripotent cells whereas during differentiation, their formation accompanies expression and assembly of lamin A/C as well as the LINC complexes on the nuclear envelope [21]. As a result, the nuclear height and shape are under their control, suggesting a role in mediating mechanosensitive processes such as motility and polarization [22].
Besides nuclear morphology, cytoplasmic forces also govern nuclear positioning in the cell by regulating the translational and rotational dynamics [23][24]. Positioning is accomplished by the physical connection by nuclear envelope proteins SUN-KASH-lma1 between centromeric heterochromatin regions and the microtubule network [25]. With the centromere providing tensional force on the microtubules that undergo dynamic instability, dynein motors mediate the rotation [26][27]. Actin links via SUN-nesprin are implicated in force transduction for nuclear movement during cell migration [28]. Regulation of nuclear position and orientation is critical in many cellular processes such as migration, cell division, polarization, fertilization and differentiation [24][26].
References
- Geiger B, Spatz JP, and Bershadsky AD. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009; 10(1):21-33. [PMID: 19197329]
- Wang N, Tytell JD, and Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009; 10(1):75-82. [PMID: 19197334]
- Geiger B, Bershadsky A, Pankov R, and Yamada KM. Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2001; 2(11):793-805. [PMID: 11715046]
- Boban M, Braun J, and Foisner R. Lamins: ‘structure goes cycling’. Biochem. Soc. Trans. 2010; 38(Pt 1):301-6. [PMID: 20074079]
- Coffinier C, Chang SY, Nobumori C, Tu Y, Farber EA, Toth JI, Fong LG, and Young SG. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc. Natl. Acad. Sci. U.S.A. 2010; 107(11):5076-81. [PMID: 20145110]
- Schirmer EC, and Foisner R. Proteins that associate with lamins: many faces, many functions. Exp. Cell Res. 2007; 313(10):2167-79. [PMID: 17451680]
- Furukawa K, Ishida K, Tsunoyama T, Toda S, Osoda S, Horigome T, Fisher PA, and Sugiyama S. A-type and B-type lamins initiate layer assembly at distinct areas of the nuclear envelope in living cells. Exp. Cell Res. 2009; 315(7):1181-9. [PMID: 19210986]
- Wagner N, and Krohne G. LEM-Domain proteins: new insights into lamin-interacting proteins. Int. Rev. Cytol. 2007; 261:1-46. [PMID: 17560279]
- Wilson KL, and Berk JM. The nuclear envelope at a glance. J. Cell. Sci. 2010; 123(Pt 12):1973-8. [PMID: 20519579]
- Dahl KN, Kahn SM, Wilson KL, and Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell. Sci. 2004; 117(Pt 20):4779-86. [PMID: 15331638]
- Ahmed S, and Brickner JH. Regulation and epigenetic control of transcription at the nuclear periphery. Trends Genet. 2007; 23(8):396-402. [PMID: 17566592]
- Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, and Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 2011; 286(30):26743-53. [PMID: 21652697]
- Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, and Lee RT. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 2005; 170(5):781-91. [PMID: 16115958]
- Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT, Wheeler MA, Ellis JA, Skepper JN, Vorgerd M, Schlotter-Weigel B, Weissberg PL, Roberts RG, Wehnert M, and Shanahan CM. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 2007; 16(23):2816-33. [PMID: 17761684]
- Alam SG, Zhang Q, Prasad N, Li Y, Chamala S, Kuchibhotla R, Kc B, Aggarwal V, Shrestha S, Jones AL, Levy SE, Roux KJ, Nickerson JA, and Lele TP. The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Sci Rep 2016; 6:38063. [PMID: 27905489]
- Towbin BD, Meister P, and Gasser SM. The nuclear envelope–a scaffold for silencing? Curr. Opin. Genet. Dev. 2009; 19(2):180-6. [PMID: 19303765]
- Ingber DE. Tensegrity-based mechanosensing from macro to micro. Prog. Biophys. Mol. Biol. 2008; 97(2-3):163-79. [PMID: 18406455]
- Mazumder A, and Shivashankar GV. Emergence of a prestressed eukaryotic nucleus during cellular differentiation and development. J R Soc Interface 2010; 7 Suppl 3:S321-30. [PMID: 20356876]
- Li Q, Makhija E, Hameed FM, and Shivashankar GV. Micropillar displacements by cell traction forces are mechanically correlated with nuclear dynamics. Biochem. Biophys. Res. Commun. 2015; 461(2):372-7. [PMID: 25911321]
- Kim D, and Wirtz D. Cytoskeletal tension induces the polarized architecture of the nucleus. Biomaterials 2015; 48:161-72. [PMID: 25701041]
- Zhong Z, Wilson KL, and Dahl KN. Beyond lamins other structural components of the nucleoskeleton. Methods Cell Biol. 2010; 98:97-119. [PMID: 20816232]
- Young KG, and Kothary R. Spectrin repeat proteins in the nucleus. Bioessays 2005; 27(2):144-52. [PMID: 15666356]
- Holaska JM, Kowalski AK, and Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004; 2(9):E231. [PMID: 15328537]
- Pederson T, and Aebi U. Actin in the nucleus: what form and what for? J. Struct. Biol. 140(1-3):3-9. [PMID: 12490148]
- Gundersen GG, and Worman HJ. Nuclear positioning. Cell 2013; 152(6):1376-89. [PMID: 23498944]
- Bustamante C, Bryant Z, and Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature 2003; 421(6921):423-7. [PMID: 12540915]
- Luger K, and Hansen JC. Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 2005; 15(2):188-96. [PMID: 15837178]
- Marenduzzo D, Micheletti C, and Cook PR. Entropy-driven genome organization. Biophys. J. 2006; 90(10):3712-21. [PMID: 16500976]


