How is SRF signaling activated?
Myocardin, MAL (or MRTF-A) and MRTF-B are the best-studied examples of MRTFs. In the cytoplasm, MRTFs exist in a stable complex with monomeric G-actin via RPEL domains, which exist at their N terminal [1]. These domains also include the Nucleus Localization Signal (NLS). The upstream regulation of MRTF-SRF activation is through Rho signaling, which is activated in response to numerous stimuli, including TGF-B, Wnt signaling, and importantly, integrin and cadherin signaling from focal adhesions and adherens junctions, respectively.
Stimulation of Rho-GTPases promotes monomeric G-actin polymerization into F-actin, liberating MRTFs from G-actin and exposing their NLS, which allows their relocation from the cytoplasm to the nucleus. For this reason, if actin polymerization is low, MRTFs are primarily found in the cytoplasm in their inactive, G-actin bound complex (with the exception of myocardin, which seems to be constitutively nuclear [1][2]).
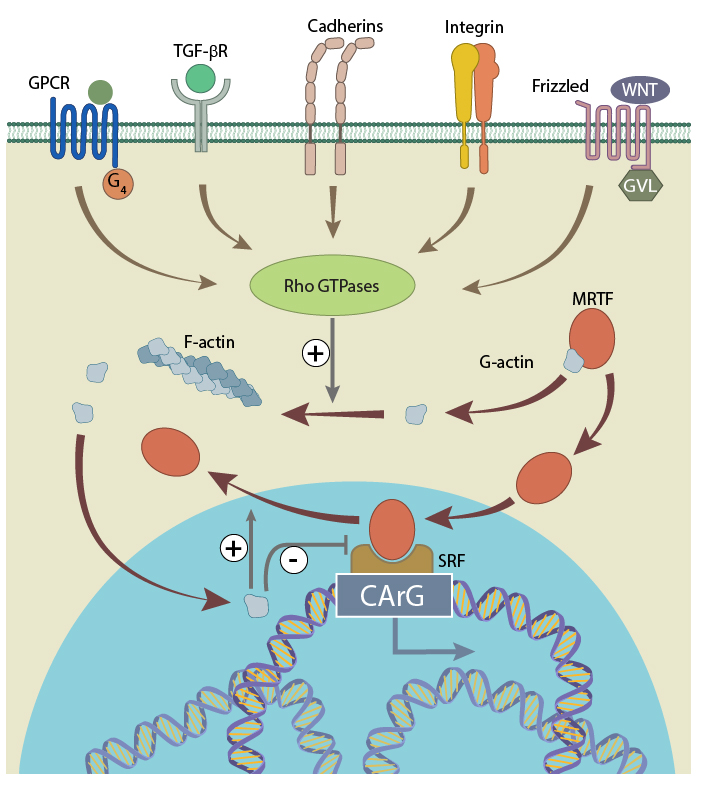
Serum response factor is activated by MRTF, which shuttles to and from the cytoplasm depending on the concentration of free G-actin (figure adapted from 20414257).
Free and activated MRTF gets transported to nucleus due to the presence of the NLS. Like in the cytoplasm, MRTF can bind to free G-actin in the nucleus, and therefore any excess nuclear G-actin can deactivate the MRTF, and promote its export back to the cytoplasm [3]. However, where MRTF remains free in the nucleus, it will associate with SRF through a basic region with adjacent Glu-rich domains [1][4]. The activated MRTF-SRF complex finally binds to CArG-box to promote the transcription of the target genes.
The described mechanism of MRTF-SRF circuit varies among different tissue and cell types. For example, in smooth, cardiac and skeletal muscle cells and in fibroblasts, the nuclear concentration of MRTFs depends on G-actin concentration, whereas in neurons there is a constantly elevated level of nuclear MRTF [4]. Additionally, there are many positive and negative regulators of the MRTF-SRF circuit. These achieve their effect via competitive binding to either MRTFs or SRF.
SRF can also initiate early response gene transcription when in complex with ternary complex factors (TCFs) Elk-1, Net and Sap-1, which belong to a subfamily of the E twenty-six (Ets) domain transcription factors. In this case a ternary nucleoprotein complex is formed when SRF binds to a TCF and the serum response element (SRE) of the c-fos promoter. This complex will again bind to target genes via the CArG-box. This G-actin-independent signaling pathway is activated by MAP kinase [5], which is controlled by upstream Ras signaling. At least four distinct MAP kinase pathways may activate TCF binding to SRF. The best known are the ERK (extracellular signal regulated kinase), JNK (c-Jun N-terminal kinase) and p38 cascades (reviewed in [6]). The ERK cascade responds to growth factors and mitogens, whereas the JNK and p38 cascades are triggered by cytokines and stress.
Other cofactors can regulate SRF. For example, the GATA, Nkx2-5 and the CRP family induce a positive regulation, whereas LIM-only protein FHL2, histone deacetylase 4, homeodomain protein MSX1 and heart-enriched homeodomain-only cofactor HOP will negatively regulate SRF activity. It is, however, not clear, to what extent these cofactors control SRF by themselves as opposed to acting in conjunction with the MRTFs or TCFs [4].
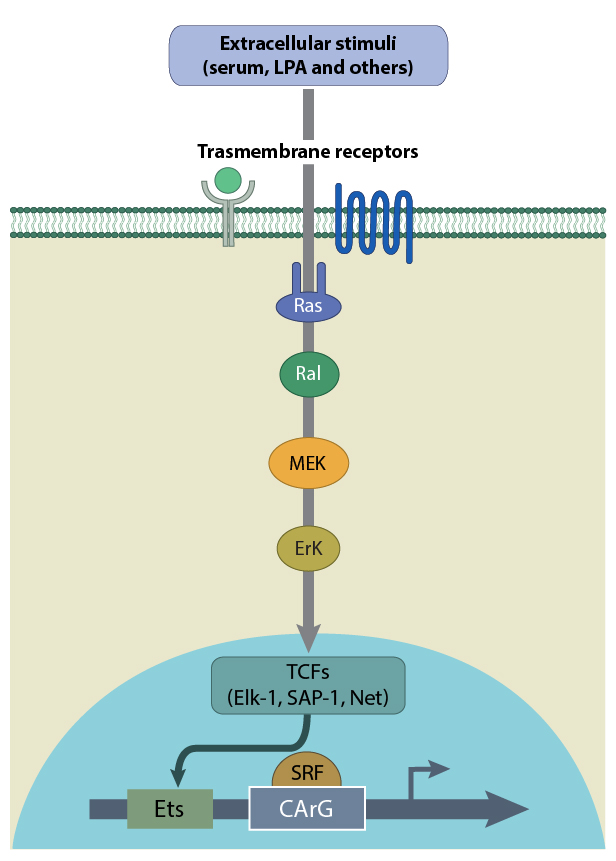
A schematic depicting the activation of SRF by TCFs such as Elk-1, SAP-1 and Net. TCFs are activated by Ras in response to extracellular stimuli.
References
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, and Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 2001; 105(7):851-62. [PMID: 11439182]
- Wang Z, Wang D, Pipes GCT, and Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. U.S.A. 2003; 100(12):7129-34. [PMID: 12756293]
- Vartiainen MK, Guettler S, Larijani B, and Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 2007; 316(5832):1749-52. [PMID: 17588931]
- Olson EN, and Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 2010; 11(5):353-65. [PMID: 20414257]
- Posern G, and Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006; 16(11):588-96. [PMID: 17035020]
- Robinson MJ, and Cobb MH. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 1997; 9(2):180-6. [PMID: 9069255]


