How is integrin activated?
Integrin activation is an important mechanism through which cells regulate integrin function by manipulating the ligand affinity of integrins spatially and temporally. Structural and functional studies suggest that integrins can exist in different ligand affinity states – low, intermediate and high (reviewed in [1]). Crystal structures have revealed that integrin heterodimers, occur in an inactive, bent V-shape with the head close to the membrane-proximal regions of the legs [2][3], maintained by the α/β salt bridge at the inner membrane region and helix packing of the transmembrane (TM) region [4]. This low affinity structure undergoes rapid, reversible conformational changes to increase ligand affinity, termed “activation” (reviewed in [5][6][7]).
The structural hallmarks of integrin activation are:
- complete extension of the extracellular domains and
- separation of the cytoplasmic leg domains (structural rearrangements detailed in [8][9]).
This process facilitates integrin-mediated signaling, thus mechano-sensing and mechano-transmitting.
Integrin can be activated from two directions, from the inside by the regulated binding of proteins to the cytoplasmic tails, and from the outside by multivalent ligand binding. In either case, talin binding to the integrin β tails is an essential and the final common step ([10], reviewed in [11]). Though the two processes are conceptually separate, they are mutually cooperative i.e one can lead to the other. Some structural studies done with force application to mimic ligand/intracellular protein suggested that the combined action of these two events favor the transition from the closed, low affinity to a open, high affinity conformation of integrin [12]. Activation leads to bidirectional signaling crucial in a variety of anchorage-dependent events such as adhesion, cell spreading, migration, polarity and organization of the ECM leading to physiological changes (reviewed in [13]). Here we discuss only well-established events that occur at the close proximity of integrin molecules localized on the plasma membrane.
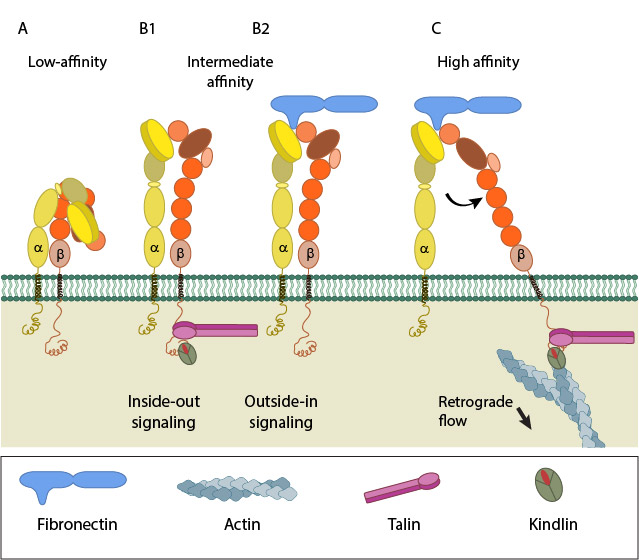
A. Low-affinity integrin has an inactive, bent, conformation. B1 and B2. Inside-out integrin activation by cytoplasmic proteins or Outside-in integrin activation via ECM ligands both lead to complete extension of the extracellular domains. C. The hallmark of open, high-affinity activated integrin is separation of the cytoplasmic leg domains.
Inside-out signaling versus Outside-in signaling
Different signaling pathways can initiate integrin activation via:
Inside-out signaling
Signals received by other receptors foster the binding of talin and kindlin to cytoplasmic end of the integrin β subunit [14], at sites of actin polymerization. Substantial information on signaling pathway leading activation is available for integrin αIIbβ3 [15].
Talin binds to integrin β-tail via F3 phospho-tyrosine binding (PTB) domain [16], a unique interaction with the membrane proximal (MP) region of the integrin (NPxY motif). This permits competition between conserved lysine on talin and an aspartic acid on integrin α essential for α/β salt bridge disruption and sufficient for integrin activation [17][18]. Addition interactions through the basic patches in the FERM subdomain F2 helps to orient the β-subunit to promote spatial separation of the cytoplasmic domains [19][20].
Kindlin is also an essential co-activator of integrin [21][22] and binds to a membrane distal NxxY motif on β-integrin via its FERM F3 subdomain [23]. A preceding threonine patch on integrins β1 and β3 that gets phosphorylated [23][24] and a tryptophan on kindlin F3 are also required for binding. However, kindlins are not known to activate integrins on their own but may render integrin-specific effects (reviewed in [25]).
The mechanism of crosstalk between integrin, talin and kindlin are not well established (reviewed in [26]). However, substantial data on the order of their binding is available. Latest Findings Talin is recruited directly to FAs from the cytosol suggesting that it does not bind to free diffusing integrins outside FAs [27] and also requires vinculin and F-actin for its activation [28]. Hence it is believed that only F-actin anchored talin at FAs bind free diffusing integrin promoting its activation [27]. Talin can directly connect to actin while kindlin links through adaptors such as migfilin, filamin, FAK,VASP and α-actinin (reviewed in [25]).
Outside-in signaling
Ligand binding to external domain causes conformational changes that increase ligand affinity, modify protein-interaction sites in the cytoplasmic domains and thence the resulting signals.
Besides conformational changes that extend integrin dimers ([29], reviewed in [13][30]), multivalent ligand binding leads to clustering of integrins, which in turn activates Src family of kinases (SFKs) by autophosphorylation [31]. SFKs phosphorylate tyrosines of the integrin cytoplasmic domain (NPxY motifs) [32][33] and other proteins [34][35] leading to:
- control of ligand binding strength
- alteration of binding with signaling molecules (kinases, GTPases and adaptors) [36], that constitute dynamic adhesion structures such as focal adhesions and podosomes (reviewed in [37][38]).
Nevertheless, whether clustering triggers outside-in signaling to facilitate integrin activation or occurs after integrin activation is uncertain (reviewed in [11][37]).
How is integrin activation regulated?
Regulation of Integrin Activation
The following classes of proteins/ events are known to enhance or inhibit integrin activation in various contexts. It must be noted that this list provides just a few examples of proteins that regulate the activation of integrin.
Regulators of talin recruitment (proteins with membrane targeting motif that bind to talin)
- Rap1-RIAM (Talin activation)
- WAVE
- Fam38A/r-Ras (Talin activation by calpain cleavage)
Phosphorylation of integrin cytoplasmic domains
- Tyr Phosphorylation -promote reduce talin affinity, promote Dok1 binding or other PTB domain proteins
- PKA mediated Ser phosphorylation- α4β1 recruitment and activation at the leading edge
- Src-kinase -mediated Tyr phosphorylation of beta integrin
Competitors for talin
- Filamin and migfilin
- CIB1 and its paralogues binding to α cytoplasmic tail
- PIPKIγ90
Others
- ERK1 and ERK2
- Trans-dominant inhibition of activation
References
- Luo B, Carman CV, and Springer TA. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007; 25:619-47. [PMID: 17201681]
- Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, and Arnaout MA. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science 2001; 294(5541):339-45. [PMID: 11546839]
- Xiong J, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, and Arnaout MA. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science 2002; 296(5565):151-5. [PMID: 11884718]
- Partridge AW, Liu S, Kim S, Bowie JU, and Ginsberg MH. Transmembrane domain helix packing stabilizes integrin alphaIIbbeta3 in the low affinity state. J. Biol. Chem. 2004; 280(8):7294-300. [PMID: 15591321]
- Shimaoka M, Takagi J, and Springer TA. Conformational regulation of integrin structure and function. Annu Rev Biophys Biomol Struct 2001; 31:485-516. [PMID: 11988479]
- Calderwood DA. Integrin activation. J. Cell. Sci. 2004; 117(Pt 5):657-66. [PMID: 14754902]
- Banno A, and Ginsberg MH. Integrin activation. Biochem. Soc. Trans. 2008; 36(Pt 2):229-34. [PMID: 18363565]
- Takagi J, Petre BM, Walz T, and Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 2002; 110(5):599-11. [PMID: 12230977]
- Puklin-Faucher E, Gao M, Schulten K, and Vogel V. How the headpiece hinge angle is opened: New insights into the dynamics of integrin activation. J. Cell Biol. 2006; 175(2):349-60. [PMID: 17060501]
- Tadokoro S, Shattil SJ, Eto K, Tai V, Liddington RC, de Pereda JM, Ginsberg MH, and Calderwood DA. Talin binding to integrin beta tails: a final common step in integrin activation. Science 2003; 302(5642):103-6. [PMID: 14526080]
- Shattil SJ, Kim C, and Ginsberg MH. The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol. 2010; 11(4):288-300. [PMID: 20308986]
- Zhu J, Luo B, Xiao T, Zhang C, Nishida N, and Springer TA. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell 2008; 32(6):849-61. [PMID: 19111664]
- Takada Y, Ye X, and Simon S. The integrins. Genome Biol. 2007; 8(5):215. [PMID: 17543136]
- Watanabe N, Bodin L, Pandey M, Krause M, Coughlin S, Boussiotis VA, Ginsberg MH, and Shattil SJ. Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin alphaIIbbeta3. J. Cell Biol. 2008; 181(7):1211-22. [PMID: 18573917]
- Han J, Lim CJ, Watanabe N, Soriani A, Ratnikov B, Calderwood DA, Puzon-McLaughlin W, Lafuente EM, Boussiotis VA, Shattil SJ, and Ginsberg MH. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr. Biol. 2006; 16(18):1796-806. [PMID: 16979556]
- Calderwood DA, Yan B, de Pereda JM, Alvarez BG, Fujioka Y, Liddington RC, and Ginsberg MH. The phosphotyrosine binding-like domain of talin activates integrins. J. Biol. Chem. 2002; 277(24):21749-58. [PMID: 11932255]
- Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, and Campbell ID. Structural basis of integrin activation by talin. Cell 2007; 128(1):171-82. [PMID: 17218263]
- Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, Lowe ED, Vakonakis I, Bate N, Critchley DR, Ginsberg MH, and Campbell ID. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 2009; 28(22):3623-32. [PMID: 19798053]
- Saltel F, Mortier E, Hytönen VP, Jacquier M, Zimmermann P, Vogel V, Liu W, and Wehrle-Haller B. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J. Cell Biol. 2009; 187(5):715-31. [PMID: 19948488]
- Kim M, Carman CV, and Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science 2003; 301(5640):1720-5. [PMID: 14500982]
- Moser M, Nieswandt B, Ussar S, Pozgajova M, and Fässler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 2008; 14(3):325-30. [PMID: 18278053]
- Pluskota E, Dowling JJ, Gordon N, Golden JA, Szpak D, West XZ, Nestor C, Ma Y, Bialkowska K, Byzova T, and Plow EF. The integrin coactivator kindlin-2 plays a critical role in angiogenesis in mice and zebrafish. Blood 2011; 117(18):4978-87. [PMID: 21378273]
- Harburger DS, Bouaouina M, and Calderwood DA. Kindlin-1 and -2 directly bind the C-terminal region of beta integrin cytoplasmic tails and exert integrin-specific activation effects. J. Biol. Chem. 2009; 284(17):11485-97. [PMID: 19240021]
- Nilsson S, Kaniowska D, Brakebusch C, Fässler R, and Johansson S. Threonine 788 in integrin subunit beta1 regulates integrin activation. Exp. Cell Res. 2006; 312(6):844-53. [PMID: 16405888]
- Karaköse E, Schiller HB, and Fässler R. The kindlins at a glance. J. Cell. Sci. 2010; 123(Pt 14):2353-6. [PMID: 20592181]
- Moser M, Legate KR, Zent R, and Fässler R. The tail of integrins, talin, and kindlins. Science 2009; 324(5929):895-9. [PMID: 19443776]
- Rossier O, Octeau V, Sibarita J, Leduc C, Tessier B, Nair D, Gatterdam V, Destaing O, Albigès-Rizo C, Tampé R, Cognet L, Choquet D, Lounis B, and Giannone G. Integrins β1 and β3 exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nat. Cell Biol. 2012; 14(10):1057-67. [PMID: 23023225]
- Banno A, Goult BT, Lee H, Bate N, Critchley DR, and Ginsberg MH. Subcellular localization of talin is regulated by inter-domain interactions. J. Biol. Chem. 2012; 287(17):13799-812. [PMID: 22351767]
- Zhu J, Carman CV, Kim M, Shimaoka M, Springer TA, and Luo B. Requirement of alpha and beta subunit transmembrane helix separation for integrin outside-in signaling. Blood 2007; 110(7):2475-83. [PMID: 17615290]
- Arnaout MA, Mahalingam B, and Xiong J. Integrin structure, allostery, and bidirectional signaling. Annu. Rev. Cell Dev. Biol. 2005; 21:381-410. [PMID: 16212500]
- Arias-Salgado EG, Lizano S, Sarkar S, Brugge JS, Ginsberg MH, and Shattil SJ. Src kinase activation by direct interaction with the integrin beta cytoplasmic domain. Proc. Natl. Acad. Sci. U.S.A. 2003; 100(23):13298-302. [PMID: 14593208]
- Law DA, DeGuzman FR, Heiser P, Ministri-Madrid K, Killeen N, and Phillips DR. Integrin cytoplasmic tyrosine motif is required for outside-in alphaIIbbeta3 signalling and platelet function. Nature 1999; 401(6755):808-11. [PMID: 10548108]
- Datta A, Huber F, and Boettiger D. Phosphorylation of beta3 integrin controls ligand binding strength. J. Biol. Chem. 2001; 277(6):3943-9. [PMID: 11723131]
- Miyamoto S, Akiyama SK, and Yamada KM. Synergistic roles for receptor occupancy and aggregation in integrin transmembrane function. Science 1995; 267(5199):883-5. [PMID: 7846531]
- Arias-Salgado EG, Lizano S, Shattil SJ, and Ginsberg MH. Specification of the direction of adhesive signaling by the integrin beta cytoplasmic domain. J. Biol. Chem. 2005; 280(33):29699-707. [PMID: 15937333]
- Anthis NJ, Haling JR, Oxley CL, Memo M, Wegener KL, Lim CJ, Ginsberg MH, and Campbell ID. Beta integrin tyrosine phosphorylation is a conserved mechanism for regulating talin-induced integrin activation. J. Biol. Chem. 2009; 284(52):36700-10. [PMID: 19843520]
- Ginsberg MH, Partridge A, and Shattil SJ. Integrin regulation. Curr. Opin. Cell Biol. 2005; 17(5):509-16. [PMID: 16099636]
- Gahmberg CG, Fagerholm SC, Nurmi SM, Chavakis T, Marchesan S, and Grönholm M. Regulation of integrin activity and signalling. Biochim. Biophys. Acta 2009; 1790(6):431-44. [PMID: 19289150]


