How does chromatin remodeling impact gene transcription?
While chromosome territory dynamics is believed to regulate gene expression through the redistribution of genes and the subsequent co-localization of these genes with transcription machinery, changes are also commonly made to the chromosome structure at a ‘local’ level. Although these changes do not necessarily involve the redistribution of genes, they do have a significant influence on gene regulation.
Post-translational modifications and biochemical remodelling of chromatin
Alterations to the chromatin structure occur as a consequence of the limitations acquired through its condensation. As chromatin is condensed into the primary nucleosome structure, DNA becomes less accessible for transcription factors. With the loosening of this chromatin structure, however, transcription machinery is better able to access the genomic DNA, and transcription is thus promoted. Hence, nucleosome organization and dynamics are regularly modified by the combined influence of covalent post-translational modifications (PTMs), histone chaperones, ATP-dependent nucleosome remodelers and histone variants [1][2][3].
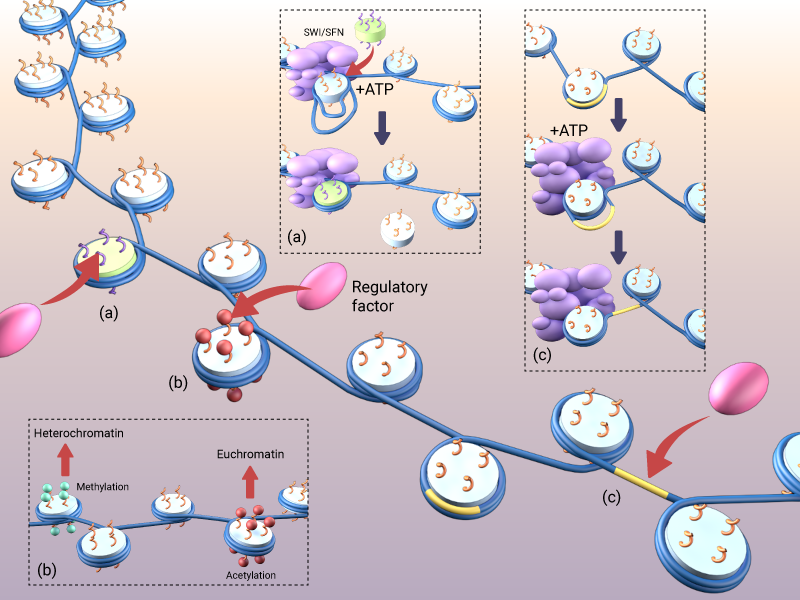
Common local changes that can be made to chromosome structures include: (a) replacement of an octamer via ATP-dependent chromatin-remodeling enzymes, (b) stabilisation or destabilisation of the chromatin via methylation and acetaylation, respectively, and (c) repositioning of nucleosomal DNA to enable the binding of a regulatory factor
To date, over 100 distinct posttranslational modifications (PTMs) of histone have been described [3]. These often take place at the N-terminal of the histone tails [3], and may include, but are not limited, to various acetylation, methylation and phosphorylation reactions. Although not all PTMs can be correlated with an increase or decrease in the expression of a given gene, several trends have been identified. Firstly, acetylation, phosphorylation and ADP-ribosylation, generally weaken charge-dependent interactions between histones and DNA, thus increasing the accessibility of genetic material to transcription machinery [4][5][6]. Lysine methylation, on the other hand, tends to increase nucleosomal stability and promotes heterochromatin formation [7], thereby reducing the accessibility of DNA. Of course, histone formation is dynamic, and the rate of histone turnover can occur rapidly [5]. It has therefore been proposed that mechanisms exist to maintain specific PTMs even in the face of ongoing nucleosome turnover and DNA replication. Although the mechanisms behind this are unclear, it has been suggested that that some histone-modifying enzymes (e.g., HDACs, methyltransferases Suv39h, SETDB1, SetD8 and G9a) remain associated with chromatin during its turnover. This would allow them to modify their cognate residues immediately following the deposition of new histones [8][9].
The chromatin structure may also be altered by ATP-dependent chromatin-remodeling enzymes which use the energy of ATP hydrolysis to mobilize nucleosomes along DNA, evict histones off DNA or promote the exchange of histone variants (reviewed in [10]). Four families of chromatin-remodeling ATPases have been described. These are classified based on their domain structures and include the SWI/SNF, (switching defective/sucrose non-fermenting) family, the ISWI (imitation SWI) family, the NuRD (nucleosome remodeling and deacetylation)/Mi-2/CHD (chromodomain, helicase, DNA binding) family and the INO80 inositol requiring 80) family [11][12].
The SWI/SFN family, and ISWI family, are the two most-studied and best-characterized classes of chromatin-remodeling complexes. Both of them can bind to either DNA or nucleosomes with various degree of affinity. After binding to their target, these complexes utilize ATP hydrolysis to destabilize histone-DNA contacts (reviewed in [13]. SWI/SNF-based remodelers presumably act by rotating DNA along its axis which subsequently generates positive supercoils. This may lead to disruption of histone octamer and transfer of histones to different DNA locations, as well as to formation of altered dimeric nucleosomes with enhanced access to DNA regulatory regions [14][15]. ISWI ATP-dependent remodeling, on the other hand, involves the formation of loops or bulges in the DNA that can propagate throughout the DNA on the octamer surface. Similarly to SWI/SNF, this ultimately leads to destabilization of octamer-DNA complexes and nucleosome remodeling [16][17]. Finally, after histone displacement, the remodeling complexes have to be released from their targets. The actual mechanism of this release and the potential role of ATP hydrolysis in this process remains poorly understood.
Physical manipulation of chromatin through mechanical force
Chromatin is physically linked to the cytoskeleton via LINC complexes that bridge the nuclear envelope. As such, chromatin is prone to local remodelling by mechanical forces generated within contractile actomyosin network. The mechanical stretching of chromatin was shown for example to directly induce the upregulation of a DHFR transgene. This was in response to the application of local stresses to the cell membrane using three-dimensional magnetic twisting cytometry. It was suggested by the authors that chromatin need only be stretched 20-100nm before it is sufficiently decondensed to allow transcription machinery to access the DHFR gene [18].
References
- Talbert PB, and Henikoff S. Histone variants–ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 2010; 11(4):264-75. [PMID: 20197778]
- Hargreaves DC, and Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 2011; 21(3):396-420. [PMID: 21358755]
- Zentner GE, and Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013; 20(3):259-66. [PMID: 23463310]
- Hong L, Schroth GP, Matthews HR, Yau P, and Bradbury EM. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J. Biol. Chem. 1993; 268(1):305-14. [PMID: 8416938]
- Banerjee T, and Chakravarti D. A peek into the complex realm of histone phosphorylation. Mol. Cell. Biol. 2011; 31(24):4858-73. [PMID: 22006017]
- Messner S, and Hottiger MO. Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol. 2011; 21(9):534-42. [PMID: 21741840]
- Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, and Workman JL. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 2012; 489(7416):452-5. [PMID: 22914091]
- Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, and Mazo A. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell 2012; 150(5):922-33. [PMID: 22921915]
- Lo SM, Follmer NE, Lengsfeld BM, Madamba EV, Seong S, Grau DJ, and Francis NJ. A bridging model for persistence of a polycomb group protein complex through DNA replication in vitro. Mol. Cell 2012; 46(6):784-96. [PMID: 22749399]
- Wang GG, Allis CD, and Chi P. Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends Mol Med 2007; 13(9):373-80. [PMID: 17822959]
- Becker PB, and Hörz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2001; 71:247-73. [PMID: 12045097]
- Bao Y, and Shen X. SnapShot: chromatin remodeling complexes. Cell 2007; 129(3):632. [PMID: 17482554]
- Johnson CN, Adkins NL, and Georgel P. Chromatin remodeling complexes: ATP-dependent machines in action. Biochem. Cell Biol. 2005; 83(4):405-17. [PMID: 16094444]
- Lorch Y, Zhang M, and Kornberg RD. RSC unravels the nucleosome. Mol. Cell 2001; 7(1):89-95. [PMID: 11172714]
- Schnitzler GR, Cheung CL, Hafner JH, Saurin AJ, Kingston RE, and Lieber CM. Direct imaging of human SWI/SNF-remodeled mono- and polynucleosomes by atomic force microscopy employing carbon nanotube tips. Mol. Cell. Biol. 2001; 21(24):8504-11. [PMID: 11713285]
- Phelan ML, Schnitzler GR, and Kingston RE. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol. Cell. Biol. 2000; 20(17):6380-9. [PMID: 10938115]
- Kassabov SR, Henry NM, Zofall M, Tsukiyama T, and Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 2002; 22(21):7524-34. [PMID: 12370299]
- Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, and Wang N. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater 2016; 15(12):1287-1296. [PMID: 27548707]


