How does Arp2/3-mediate the nucleation of branched filaments?
The Arp2/3 complex is composed of 7 evolutionarily conserved subunits (Arp2, Arp3, ARPC1-C5) that are structurally similar to the barbed end of actin [1]. The complex is inherently inactive, however once activated it facilitates the nucleation of actin monomers from existing filaments as new branches or daughter filaments (reviewed in [2]).
The Arp2/3 complex is essential in the formation of lamellipodia, where it facilitates nucleation exclusively at the leading edge (as opposed to regions further away from the front). Although it is absent from the shafts of filopodia [3], it is known to play a critical role in initiation and growth of these structures [4]. Conversely, the Arp2/3 complex is present along the length of shafts of invadopodia. It has also been detected at the base of invadopodia, whilst being absent from the tips [5].
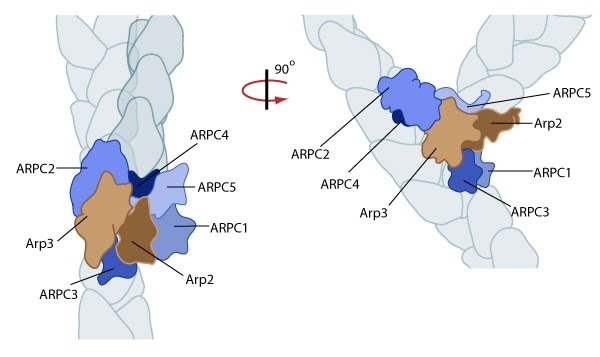
Arp2/3 complex initiates a new branched filament by binding to the side of the mother filament and recruiting actin monomers. Components of the Arp2/3 complex remain at the pointed end of the filament (Figure adapted from [18316411]).
The exact mechanism of nucleation remains unclear however it is believed that all seven subunits coordinate to anchor the pointed end of the new filament at a 78° angle to the existing actin network [10]. NPFs act to deliver actin subunits to the Arp2/3 complex at the barbed end, via binding through their WH2 domain – this serves to push the leading edge of a cell forward [11]. It has also been suggested that each component of the complex plays specific roles, with Arp2 and Arp3 interacting with the pointed end of the daughter filament and ARPC2 and ARPC4 binding to the original filament [10].
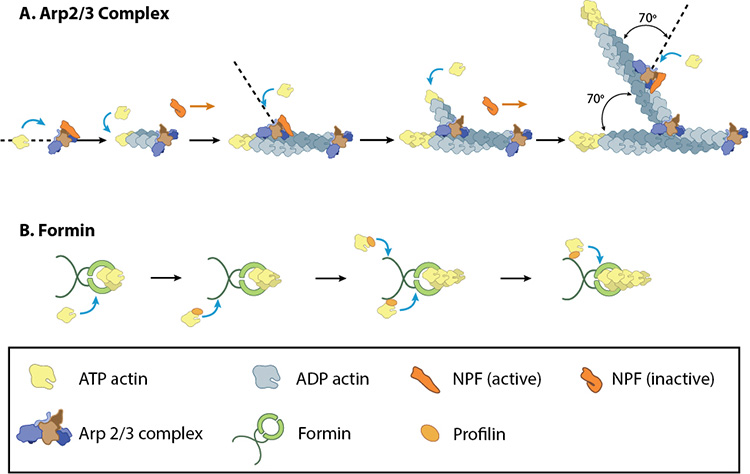
A: NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.
Importantly, the compressive stresses placed upon actin filaments may cause them to bend and curve. This property of the actin cytoskeleton was shown to regulate Arp2/3 mediated nucleation [12]. Here, a significantly higher frequency of filament branches were observed originating from the convex side of curved filaments compared to the concave side. This indicated that nucleation was more likely to occur if an extensional strain existed on the Arp2/3 binding surface on the filament compared to a compressional strain, which generates the concave surface [12].
This bias of Arp2/3 towards branch formation on convex curvature was proposed to represent a mechanosensing mechanism whereby detection of filament curvature alters the local cytoskeleton in order to reinforce it against the imposing forces [12].
What are tandem-monomer-binding nucleators?
Although the most commonly described nucleators are the Arp2/3 complex, and the formins, a third group, known as ‘tandem-monomer-binding nucleators’, has also been identified. Each member possesses tandem repeats of G-actin binding motifs.
Included in this group of nucleators are the Spire proteins, Cordon-bleu (Cobl), Leiomodin (Lmod-2), JMY and adenomatous polyposis coli (APC). Common to each of these proteins are repeats of the actin binding motif Wiskott-Aldrich Syndrome protein (WASp) homology 2 (WH2) domain. However, additional actin binding motifs may be present in the individual members, providing variation not their mechanisms of nucleation, but importantly, in the cellular functions they facilitate.
References
- Beltzner CC, and Pollard TD. Identification of functionally important residues of Arp2/3 complex by analysis of homology models from diverse species. J. Mol. Biol. 2004; 336(2):551-65. [PMID: 14757065]
- Firat-Karalar EN, and Welch MD. New mechanisms and functions of actin nucleation. Curr. Opin. Cell Biol. 2010; 23(1):4-13. [PMID: 21093244]
- Svitkina TM, and Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 1999; 145(5):1009-26. [PMID: 10352018]
- Korobova F, and Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol. Biol. Cell 2008; 19(4):1561-74. [PMID: 18256280]
- Schoumacher M, Goldman RD, Louvard D, and Vignjevic DM. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 2010; 189(3):541-56. [PMID: 20421424]
- Xu J, Wang F, Van Keymeulen A, Herzmark P, Straight A, Kelly K, Takuwa Y, Sugimoto N, Mitchison T, and Bourne HR. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell 2003; 114(2):201-14. [PMID: 12887922]
- Suetsugu S, Kurisu S, Oikawa T, Yamazaki D, Oda A, and Takenawa T. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J. Cell Biol. 2006; 173(4):571-85. [PMID: 16702231]
- Lorenz M, Yamaguchi H, Wang Y, Singer RH, and Condeelis J. Imaging sites of N-wasp activity in lamellipodia and invadopodia of carcinoma cells. Curr. Biol. 2004; 14(8):697-703. [PMID: 15084285]
- Stradal TEB, Pusch R, and Kliche S. Molecular regulation of cytoskeletal rearrangements during T cell signalling. Results Probl Cell Differ 2006; 43:219-44. [PMID: 17068974]
- Rouiller I, Xu X, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, and Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 2008; 180(5):887-95. [PMID: 18316411]
- Pollard TD, and Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003; 112(4):453-65. [PMID: 12600310]
- Risca VI, Wang EB, Chaudhuri O, Chia JJ, Geissler PL, and Fletcher DA. Actin filament curvature biases branching direction. Proc. Natl. Acad. Sci. U.S.A. 2012; 109(8):2913-8. [PMID: 22308368]


