How do actin filaments grow?
Actin Filaments (F-actin) grow from the polymerization of G-actin monomers
Actin is a highly abundant (10-100 micromolar on average),~42 kDa structural protein found in all eukaryotic cells (except for nematode sperm). With more than 95% conservation in the primary structure, actin is one of the most highly-conserved proteins [1]. The monomeric, globular form of actin, known as G-actin, forms the basic unit for actin filaments. In many cases actin filaments may bundle together with other actin filaments, or, together with their associated motor proteins (e.g. myosin superfamily) form an elaborate network known as the actin cytoskeleton. This occurs primarily at or near the plasma membrane. Consequently a region of high actin filament density is commonly found at the cell periphery and is known as the cell cortex. Actin filaments in the cell cortex determine the shape, stiffness and movement of the cell surface. The actin cytoskeleton also facilitates the transduction of mechanical signals, and can generate the intracellular forces that are required for many cellular functions including cell motility, muscle contraction, cell division, cytokinesis, vesicle and organelle movement and cell signaling. Actin also contributes to the formation and maintenance of cell junctions.
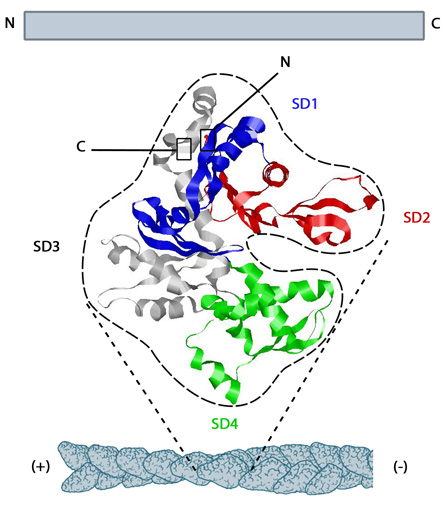
The structure shown here [3] was downloaded from the RSCB Protein Data Bank (PDB file: 1atn). The ATP binding cleft is starred (*) on the right. Actin comprises four subdomains, termed SD1 (blue), SD2 (red), SD3 (grey) and SD4 (green). The barbed end of each monomer (SD1 and SD3) is shown on the left and the pointed end (SD2 and SD4) is shown on the right. Similarly the polarity of the actin filament, which comprises these monomers is shown at the bottom, with the barbed (+) end on the left and the pointed (-) end on the right.
How do actin monomers polymerize to form an actin filament?
Actin filaments are highly dynamic and their polymerization is usually correlated to their disassembly. Generally, actin filament polymerization occurs over three phases: A nucleation phase, an elongation phase and a steady state phase.
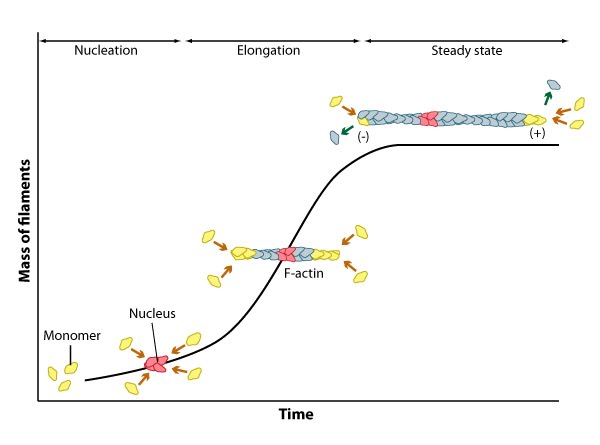
Nucleation, elongation, and steady state phase of actin filament assembly.
During the nucleation phase the formation of a stable ‘actin nucleus’ occurs. This is usually comprised of three actin monomers in complex. In the elongation phase monomers are rapidly added to the filament at the (+ve) or barbed end and this is often facilitated by additional elongation factors such as formin. For this process to occur, the (+) end of the filament must be exposed, and this means removal of capping protein.
References
- Elzinga M, Collins JH, Kuehl WM, and Adelstein RS. Complete amino-acid sequence of actin of rabbit skeletal muscle. Proc. Natl. Acad. Sci. U.S.A. 1973; 70(9):2687-91. [PMID: 4517681]
- Holmes KC, Popp D, Gebhard W, and Kabsch W. Atomic model of the actin filament. Nature 1990; 347(6288):44-9. [PMID: 2395461]
- Egelman EH. The structure of F-actin. J. Muscle Res. Cell. Motil. 1985; 6(2):129-51. [PMID: 3897278]
- Orlova A, Galkin VE, VanLoock MS, Kim E, Shvetsov A, Reisler E, and Egelman EH. Probing the structure of F-actin: cross-links constrain atomic models and modify actin dynamics. J. Mol. Biol. 2001; 312(1):95-106. [PMID: 11545588]
- Oda T, Stegmann H, Schröder RR, Namba K, and Maéda Y. Modeling of the F-actin structure. Adv. Exp. Med. Biol. 2007; 592:385-401. [PMID: 17278381]
- Aebi U, Fowler WE, Isenberg G, Pollard TD, and Smith PR. Crystalline actin sheets: their structure and polymorphism. J. Cell Biol. 1981; 91(2 Pt 1):340-51. [PMID: 7309785]
- Egelman EH, Francis N, and DeRosier DJ. F-actin is a helix with a random variable twist. Nature 1982; 298(5870):131-5. [PMID: 7201078]


