How do actin filaments depolymerize?
Introduction to Actin Filament Depolymerization
Whole cell motility and mechanosensing rely on the continual restructuring of the cytoskeleton, particularly within lamellipodia and filopodia; two dynamic structures that contribute to cell motility. Actin filament depolymerization ensures the turnover of actin filaments within these structures and maintains a pool of actin monomers that permits the continual restructuring and growth of the actin cytoskeleton.
Disassembly of actin filaments occurs at the pointed end of the filament and is driven by the ADF/cofilin (AC) family of proteins. Actin monomers intrinsically dissociate from the barbed end at a faster rate than they do from the pointed end [1]. This is counteracted by the binding of capping proteins or formins to the barbed end, creating a more stable filament. The action of cofilin at the pointed end serves to destabilize the filament and promote the release of ADP-actin monomers.
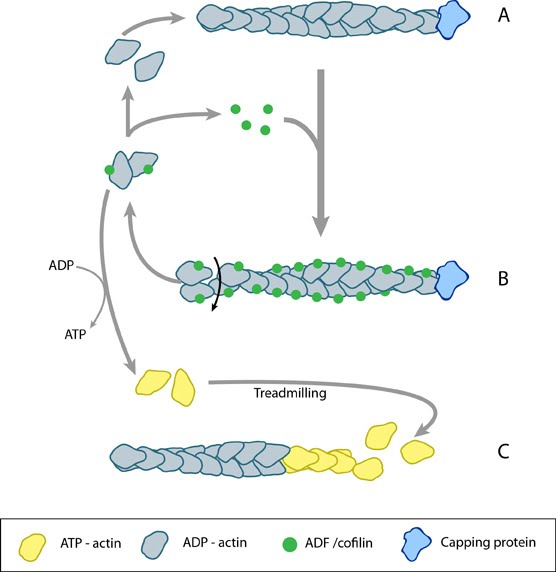
ADF/cofilin influences actin filament turnover. ADF cooperatively binds to F-actin to increase the steady state turn-over (e.g. treadmilling rate) of actin filaments and accumulation of ADF-ADP-G-actin, ADP-G-actin, and ATP-G-actin (via nucleotide exchange). ADF binds faster to actin filaments that have barbed end capping proteins (e.g. gelsolin) (B). The actin subunits depolymerizing from ADF-bound filaments are recycled to filaments lacking ADF, thus maintaining the pool of actin-filaments. At high ADF concentration, polymerization of ADP-actin at the pointed end is favored on capped-filaments (A); alternatively, nucleotide exchange increases the pool of ATP-actin for barbed end assembly of new or existing filaments (C).
The conversion of ATP-F-actin to ADP-F-actin involves the hydrolysis of ATP and subsequent release of free inorganic phosphate (Pi) molecules. These free Pi molecules bind antagonistically to cofilin and as such cofilin binding to F-actin precedes Pi release [2]. Whilst Pi stabilizes the actin filament once bound, AC proteins destabilize the filament by inducing conformational change. This involves the production or stabilization of a twist within the filament or between monomers, creating a strain that leads to the loss of filament integrity and disassembly.
The conformational change induced by cofilin binding further promotes filament destabilization through increasing the rate of Pi release by approximately 10-fold [2]. The pH of the environment also affects the ability of cofilin to depolymerize actin filaments, with a higher pH favoring depolymerization due to the weaker binding of Pi in more alkali environments [3].
Recent data suggests that the efficacy of cofilin in filament severing is enhanced in actin bundles despite slower binding kinetics of cofilin to fascin-bundled actin filaments. This enhanced efficacy permits the the severing of bundled actin filaments at concentrations of cofilin that are inadequate to induce severing of free filaments. It was proposed that this is the result of cross-linkers such as fascin reducing the flexibility of the actin filaments and subsequently making them more vulnerable to the twisting effect of cofilin; as the filament as a whole is unable to provide slack for the sections under pressure [4].
The destabilized form of actin filaments, which has been compared to that observed in younger filaments [5], is more prone to filament severing. The higher affinity of ACs for ADP-F-actin relative to ATP-F-actin causes severing in the central regions of filaments where ADP-actin is enriched, though depolymerization at the pointed end also occurs [6].
ADF / Cofilin in actin filament deploymerization
The AC protein family includes actin depolymerizing factor (ADF) [7] aka destrin (destroys F-actin)[8][3], actophorin (found in Acanthamoeba castellanii [9] and depactin (found in starfish oocytes [10]). Both the phosphorylation of ACs [11] and the membrane lipids, phosphatidyl 4-phosphate (PIP) and 4,5-bis-phosphate (PIP2) [12], greatly reduce the F-actin binding and depolymerizing activity of ACs. AC phosphorylation in vertebrates is controlled by the activity of *Rho GTPase and Lim kinase pathways [13][14]. ADF/cofilin contribute to actin dynamics in a number of cell structures including the lamellipodium, filopodium, the postsynaptic density in dendritic spines [15] and invadopodia [16].
Mechanistically, cofilin binds between actin subunits when a longitudinal bond spontaneously breaks as the filament bends in thermal motion [16]. Cooperative binding of ADF/cofilin causes the filament to twist and structurally weaken [17]; this causes a modest severing effect that results in pointed end depolymerization and a 2-3 fold decrease in the average length [6][18]. ACs promote shortening of actin filaments in two main ways: by severing the filament to create more ends that disassemble [19][20][21] and by promoting subunit loss from filament ends [6][22] [23].
ACs promote filament turnover by altering the kinetics of filament disassembly. Recent findings have found that cofilin is more efficacious at severing actin filaments that are are cross-linked in actin bundles [4]. This is believed to result from the filament being more susceptible to the cofilin-induced twist, as its rigidity is maintained by the cross-linkers and it is unable flex under the added tension.
ACs bind both F and G actin-ADP with greater avidity than they bind to F or G actin-ATP and ADF-actin-ADP has a higher dissociation rate than actin-ADP [6][20]. ACs also promote debranching by accelerating the rate-limiting release of the γ-phosphate from ADP-Pi-actin filament subunits [24]. Lastly, ACs interact with the Arp2/3 complex and ADP-actin subunits at the free pointed ends to aid in disassembly [25].
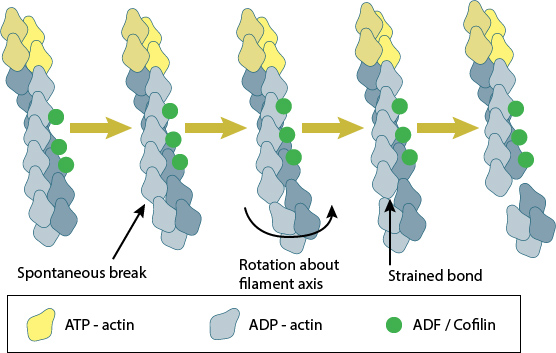
Cofilin binds between actin subunits when a longitudinal bond spontaneously breaks as the filament bends in thermal motion. Cooperative binding of ADF/cofilin causes the filament to twist and structurally weaken, resulting in severing of the actin filament.
Interestingly, the nucleation of ADP-actin is greatly facilitated by high ADF concentrations; the ADF-ADP-G-actin complex is stabilzed, the treadmilling rate declines, and pointed-end assembly of ADP-G-actin is favored due to lateral stability of actin-filaments [26]. Increased lateral stability of the filaments may form a selection process that stabilizes filaments in bundles and may account for the emergence and extension of actin-based structures such as filopodia [27]. Further support for the role of ADF/cofilin in the stabilization of actin filaments comes from invadopodia, where the loss of cofilin results in a decrease in invadopodia lifetime [28].
It should be noted that the AC proteins have been shown to initiate nucleation of new filaments from recently disassembled monomers. In this case, cofilin shows a higher nucleating activity (double) compared to ADF [3]. Based on this finding, which was observed at pH8 in experimental conditions, ADF is considered to be more effective in promoting actin turnover as the free monomers are less likely to undergo ADF mediated nucleation immediately following disassembly [3].
Actin filaments may be severed at Arp2/3 junctions
Along with the cofilin mediated severing of actin filaments, actin networks may be disassembled through a process known as F-actin debranching, which is predominately mediated by a protein known as glia maturation factor (GMF). This cofilin homolog binds to the Arp2/3 complex rather than the actin filaments themselves, severs the filaments at the branch junction and inhibits further actin filament nucleation at the site [29]. GMF mediated actin filament debranching was shown to be particularly important in lamellipodia growth, and cell migration [30]. Although less is known about the mechanism behind GMF debranching, it has been proposed to be related to the severing activity of cofilin, and may result from the partial displacement of an actin monomer of the daughter filament from the Arp2/3 complex, which occurs upon GMF binding to Arp 2 and ARPC1/p40 [31],[32].
Actin filament disassembly regulates actomyosin contraction
Along with actin filament disassembly or severing, ADF/cofilin was recently shown to carry out another important role; specifically the regulation of Myosin II mediated contractility and actomyosin formation. This was proposed to result from competitive antagonism, where myosin II must compete with cofilin for binding sites on F-actin [33].
In this study it was shown that the binding affinities of each protein are ATP dependent, with ADF/cofilin possessing a competitive advantage at cellular levels of ATP, whilst in the absence of ATP the binding affinities of each protein is similar. Importantly, a reduction in the levels of both ADF and cofilin lead to an increase in the concentration of F-actin, a finding that was attributed not to a loss in cofilin mediated F-actin severing, but rather to an increase in myosin-II dependent actin assembly via its crosslinking properties. This was confirmed with the introduction of blebbistatin which inhibited myosin II activity and subsequently lead to the disassembly of F-actin [33].
The implications for this role of ADF/cofilin may be described at the molecular level, however as shown by Wiggan O et al the consequences are clearly evident at a cellular level, with persistent membrane blebs being observed in HeLa cells depleted of the proteins [33]. As it had previously been reported that non-apoptotic blebs were produced as a means of releasing cell tension, it is probable that the observed phenotype occurred for a similar purpose, and highlights the importance of ADF/cofilin in the regulation of cortical tension and actomyosin activity [33].
Despite these findings, in some situations a cooperative relationship between ADF/cofilin and Myosin-II appears to exist. This has been described, for example, in a study investigating actomyosin ring constriction in budding yeast cells [34]. Supporting earlier findings [35][36], this study also confirmed that deletion or inhibition of the motor-domain of Myosin II (MyoI) did not completely prevent constriction, but noted that a 40% reduction in the rate of contraction was observed. This was in contrast to mutations in cofilin, or stabilization of actin filaments, which did prevent actomyosin ring constriction. Model simulations using this data indicated a role of Myosin-II in the promotion of cofilin mediated depolymerization, and it was suggested that it is the disassembly of F-actin that is the primary contributor to actomyosin ring constriction [34].
References
- Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 1986; 103(6 Pt 2):2747-54. [PMID: 3793756]
- Muhlrad A, Pavlov D, Peyser YM, and Reisler E. Inorganic phosphate regulates the binding of cofilin to actin filaments. FEBS J. 2006; 273(7):1488-96. [PMID: 16689934]
- Yeoh S, Pope B, Mannherz HG, and Weeds A. Determining the differences in actin binding by human ADF and cofilin. J. Mol. Biol. 2002; 315(4):911-25. [PMID: 11812157]
- Breitsprecher D, Koestler SA, Chizhov I, Nemethova M, Mueller J, Goode BL, Small JV, Rottner K, and Faix J. Cofilin cooperates with fascin to disassemble filopodial actin filaments. J. Cell. Sci. 2011; 124(Pt 19):3305-18. [PMID: 21940796]
- Bernstein BW, and Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010; 20(4):187-95. [PMID: 20133134]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, and Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 1997; 136(6):1307-22. [PMID: 9087445]
- Abe H, Endo T, Yamamoto K, and Obinata T. Sequence of cDNAs encoding actin depolymerizing factor and cofilin of embryonic chicken skeletal muscle: two functionally distinct actin-regulatory proteins exhibit high structural homology. Biochemistry 1990; 29(32):7420-5. [PMID: 1699599]
- Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 1999; 15:185-230. [PMID: 10611961]
- Cooper JA, and Schafer DA. Control of actin assembly and disassembly at filament ends. Curr. Opin. Cell Biol. 2000; 12(1):97-103. [PMID: 10679358]
- Mabuchi I. An actin-depolymerizing protein (depactin) from starfish oocytes: properties and interaction with actin. J. Cell Biol. 1983; 97(5 Pt 1):1612-21. [PMID: 6226671]
- Morgan TE, Lockerbie RO, Minamide LS, Browning MD, and Bamburg JR. Isolation and characterization of a regulated form of actin depolymerizing factor. J. Cell Biol. 1993; 122(3):623-33. [PMID: 7687605]
- Yonezawa N, Nishida E, Iida K, Yahara I, and Sakai H. Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease I with actin by phosphoinositides. J. Biol. Chem. 1990; 265(15):8382-6. [PMID: 2160454]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, and Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 1998; 393(6687):805-9. [PMID: 9655397]
- Edwards DC, Sanders LC, Bokoch GM, and Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1999; 1(5):253-9. [PMID: 10559936]
- Racz B, and Weinberg RJ. Spatial organization of cofilin in dendritic spines. Neuroscience 2006; 138(2):447-56. [PMID: 16388910]
- Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, and Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997; 16(18):5520-30. [PMID: 9312011]
- McGough A, Pope B, Chiu W, and Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J. Cell Biol. 1997; 138(4):771-81. [PMID: 9265645]
- Maciver SK, Zot HG, and Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J. Cell Biol. 1991; 115(6):1611-20. [PMID: 1757465]
- Cooper JA, Blum JD, Williams RC, and Pollard TD. Purification and characterization of actophorin, a new 15,000-dalton actin-binding protein from Acanthamoeba castellanii. J. Biol. Chem. 1986; 261(1):477-85. [PMID: 3941084]
- Maciver SK, Pope BJ, Whytock S, and Weeds AG. The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur. J. Biochem. 1998; 256(2):388-97. [PMID: 9760179]
- Du J, and Frieden C. Kinetic studies on the effect of yeast cofilin on yeast actin polymerization. Biochemistry 1998; 37(38):13276-84. [PMID: 9748335]
- Ressad F, Didry D, Egile C, Pantaloni D, and Carlier MF. Control of actin filament length and turnover by actin depolymerizing factor (ADF/cofilin) in the presence of capping proteins and ARP2/3 complex. J. Biol. Chem. 1999; 274(30):20970-6. [PMID: 10409644]
- Didry D, Carlier MF, and Pantaloni D. Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 1998; 273(40):25602-11. [PMID: 9748225]
- Blanchoin L, and Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 1999; 274(22):15538-46. [PMID: 10336448]
- Blanchoin L, Pollard TD, and Mullins RD. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 2000; 10(20):1273-82. [PMID: 11069108]
- Andrianantoandro E, and Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 2006; 24(1):13-23. [PMID: 17018289]
- Michelot A, Berro J, Guérin C, Boujemaa-Paterski R, Staiger CJ, Martiel J, and Blanchoin L. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr. Biol. 2007; 17(10):825-33. [PMID: 17493813]
- Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, and Condeelis J. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol. 2005; 168(3):441-52. [PMID: 15684033]
- Gandhi M, Smith BA, Bovellan M, Paavilainen V, Daugherty-Clarke K, Gelles J, Lappalainen P, and Goode BL. GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr. Biol. 2010; 20(9):861-7. [PMID: 20362448]
- Poukkula M, Hakala M, Pentinmikko N, Sweeney MO, Jansen S, Mattila J, Hietakangas V, Goode BL, and Lappalainen P. GMF promotes leading-edge dynamics and collective cell migration in vivo. Curr. Biol. 2014; 24(21):2533-40. [PMID: 25308079]
- Ydenberg CA, Padrick SB, Sweeney MO, Gandhi M, Sokolova O, and Goode BL. GMF severs actin-Arp2/3 complex branch junctions by a cofilin-like mechanism. Curr. Biol. 2013; 23(12):1037-45. [PMID: 23727094]
- Luan Q, and Nolen BJ. Structural basis for regulation of Arp2/3 complex by GMF. Nat. Struct. Mol. Biol. 2013; 20(9):1062-8. [PMID: 23893131]
- Wiggan O, Shaw AE, DeLuca JG, and Bamburg JR. ADF/cofilin regulates actomyosin assembly through competitive inhibition of myosin II binding to F-actin. Dev. Cell 2012; 22(3):530-43. [PMID: 22421043]
- Mendes Pinto I, Rubinstein B, Kucharavy A, Unruh JR, and Li R. Actin depolymerization drives actomyosin ring contraction during budding yeast cytokinesis. Dev. Cell 2012; 22(6):1247-60. [PMID: 22698284]
- Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, Iwase M, Vallen EA, and Bi E. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J. Cell Biol. 2010; 191(7):1333-50. [PMID: 21173112]
- Lord M, Laves E, and Pollard TD. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol. Biol. Cell 2005; 16(11):5346-55. [PMID: 16148042]


