Filamin
The filamin family of proteins bind to both actin and a number of signaling molecules including Rho GTPases. Evidence for this was shown with the loss of Filamin-A in M2 Melanoma cells, which prevented RalA- and Cdc42- mediated filopodia formation [1]. Filamin A is necessary for the successful production of lamellipodia [2][3], specifically due to its F-actin crosslinking activity [3]. Following recruitment to the membrane at the leading edge of a lamellipodium, it crosslinks newly polymerized actin filaments to enhance lamellipodium formation [3].
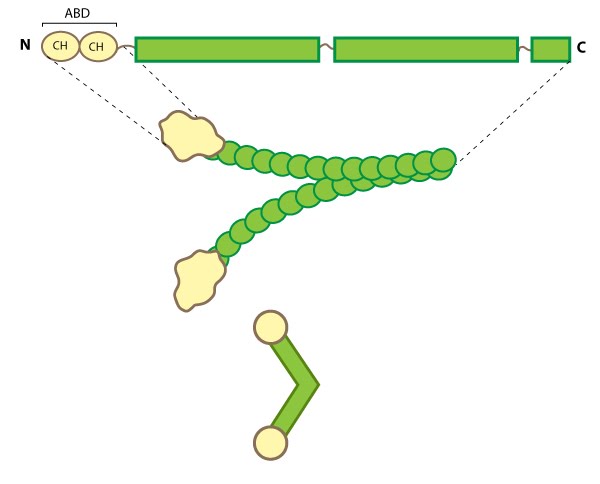
This schematic diagram illustrates the molecular organization of filamin [2391361] and provides examples for how the filamin dimer is represented in figures throughout this resource. The ABD is at the amino (N)-terminus in contrast to the opposite end (the carboxy [C]-terminus), which contains a significant number of protein-protein interaction domains (reviewed in [PMID 15516996]).
Filamin binds all actin isoforms (e.g. F-actin, G-actin) and its structural organization allows it to form a flexible bridge between two actin filaments at various angles, thereby imparting the actin network with loose or gel-like qualities [7]. Filamin also simultaneously interacts with and influences the activity of a number of other diverse proteins (e.g. transmembrane receptors, cell adhesion molecules, signaling molecules) through its immunoglobulin-like domains (reviewed in [1][8][9]). In some instances, the majority of these repeats are utilized for protein-protein interactions (e.g. beta-integrin binding [10]).
Filamin Localization and Function
Filamin forms a vital scaffolding adaptor and regulatory component that contributes to the mechanical stability of cells by linking the internal actin network with membrane receptors and mechanosensitive components. This function correlates with its distribution in cultured cells along actin stress fibers, within cortical actin networks and sometimes at membrane ruffles [11][12].
Numerous filamin isoforms exist in metazoan cells (reviewed in [1][8]) and in some cases their cellular distribution suggests that certain isoforms may have specialized roles within the cell [13]. In muscle cells, filamin is locally concentrated at specific points called Z-lines, but in non-muscle cells filamin generally remains distributed throughout larger actin-based structures such as the lamellipodium, cortical actin, stress fibers and at sites of adhesion [reviewed in [14]].
Filamin not only binds to membrane receptors and to the Rho family of GTPases to influence actin organization (reviewed in [8]), but can itself directly transduce mechanical signals (e.g. stress) to regulate the cell stiffness [15] and possibly alter cellular adhesion [16]. Filamin can be phosphorylated by numerous kinases including cAMP-dependent protein kinase (i.e. protein kinase A) [17][18] and protein kinase C [19]. Filamin phosphorylation not only regulates its interaction with other proteins and affects its ability to cross-link actin, but also regulates its stability and proteolysis (destruction) (reviewed in [1][8]).
References
- van der Flier A, and Sonnenberg A. Structural and functional aspects of filamins. Biochim. Biophys. Acta 2001; 1538(2-3):99-117. [PMID: 11336782]
- Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, and Stossel TP. Actin-binding protein requirement for cortical stability and efficient locomotion. Science 1992; 255(5042):325-7. [PMID: 1549777]
- Takabayashi T, Xie M, Takeuchi S, Kawasaki M, Yagi H, Okamoto M, Tariqur RM, Malik F, Kuroda K, Kubota C, Fujieda S, Nagano T, and Sato M. LL5beta directs the translocation of filamin A and SHIP2 to sites of phosphatidylinositol 3,4,5-triphosphate (PtdIns(3,4,5)P3) accumulation, and PtdIns(3,4,5)P3 localization is mutually modified by co-recruited SHIP2. J. Biol. Chem. 2010; 285(21):16155-65. [PMID: 20236936]
- Wang K, Ash JF, and Singer SJ. Filamin, a new high-molecular-weight protein found in smooth muscle and non-muscle cells. Proc. Natl. Acad. Sci. U.S.A. 1975; 72(11):4483-6. [PMID: 53835]
- Shizuta Y, Shizuta H, Gallo M, Davies P, and Pastan I. Purification and properties of filamin, and actin binding protein from chicken gizzard. J. Biol. Chem. 1976; 251(21):6562-7. [PMID: 977588]
- Hock RS, Davis G, and Speicher DW. Purification of human smooth muscle filamin and characterization of structural domains and functional sites. Biochemistry 1990; 29(40):9441-51. [PMID: 2248958]
- Gardel ML, Nakamura F, Hartwig JH, Crocker JC, Stossel TP, and Weitz DA. Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc. Natl. Acad. Sci. U.S.A. 2006; 103(6):1762-7. [PMID: 16446458]
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, and Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001; 2(2):138-45. [PMID: 11252955]
- Feng Y, and Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat. Cell Biol. 2004; 6(11):1034-8. [PMID: 15516996]
- Loo DT, Kanner SB, and Aruffo A. Filamin binds to the cytoplasmic domain of the beta1-integrin. Identification of amino acids responsible for this interaction. J. Biol. Chem. 1998; 273(36):23304-12. [PMID: 9722563]
- Chiang W, Greaser ML, and Lyons GE. Filamin isogene expression during mouse myogenesis. Dev. Dyn. 2000; 217(1):99-108. [PMID: 10679933]
- van der Ven PF, Obermann WM, Lemke B, Gautel M, Weber K, and Fürst DO. Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil. Cytoskeleton 2000; 45(2):149-62. [PMID: 10658210]
- Pavalko FM, Otey CA, and Burridge K. Identification of a filamin isoform enriched at the ends of stress fibers in chicken embryo fibroblasts. J. Cell. Sci. 1989; 94 ( Pt 1):109-18. [PMID: 2693470]
- Coleman TR, and Mooseker MS. Effects of actin filament cross-linking and filament length on actin-myosin interaction. J. Cell Biol. 1985; 101(5 Pt 1):1850-7. [PMID: 2932451]
- Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, and McCulloch CA. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J. Biol. Chem. 1998; 273(3):1689-98. [PMID: 9430714]
- Kim H, Sengupta A, Glogauer M, and McCulloch CA. Filamin A regulates cell spreading and survival via beta1 integrins. Exp. Cell Res. 2007; 314(4):834-46. [PMID: 18177638]
- Wallach D, Davies PJ, and Pastan I. Purification of mammalian filamin. Similarity to high molecular weight actin-binding protein in macrophages, platelets, fibroblasts, and other tissues. J. Biol. Chem. 1978; 253(9):3228-35. [PMID: 641065]
- Wallach D, Davies PJ, and Pastan I. Cyclic AMP-dependent phosphorylation of filamin in mammalian smooth muscle. J. Biol. Chem. 1978; 253(13):4739-45. [PMID: 207708]
- Kawamoto S, and Hidaka H. Ca2+-activated, phospholipid-dependent protein kinase catalyzes the phosphorylation of actin-binding proteins. Biochem. Biophys. Res. Commun. 1984; 118(3):736-42. [PMID: 6231024]


